Wastewater-based epidemiology indirectly monitors infection rates in a community based on pathogen levels in local wastewater. Because symptomatic and asymptomatic SARS-CoV-2 infected individuals shed viral RNA in sewer systems, analysis of SARS-CoV-2 in sewage networks can provide information about SARS-CoV-2 infections within the community. However, the accuracy of these data depends on the quality of the wastewater testing method. During the COVID-19 pandemic, Droplet Digital PCR has proven that it has the sensitivity needed to detect outbreaks and monitor SARS-CoV-2 variants in communities via wastewater testing.
COVID-19 Poses Persistent Public Health Challenges
ddPCR Testing for Wastewater Surveillance
ddPCR technology uses a water-emulsion droplet system to partition nucleic acid samples and perform PCR amplification within each droplet. Droplet Digital PCR can be used to detect SARS-CoV-2 viral RNA shed in wastewater, providing real-time surveillance of spread at the community level. In a recent study, fluctuations in SARS-CoV-2 wastewater measurements predicted increases in confirmed case numbers in the area (Gonzalez et al. 2020).
Key Droplet Digital PCR facts:
- Compliant with U.S. Food and Drug Administration (FDA) guidance for
SARS-CoV-2 quantification - Recommended by the U.S. Centers for Disease Control (CDC) over bulk quantification methods
- Published as a dependable viral quantification method for wastewater testing by the U.S. Environmental Protection Agency (EPA) (Jahne et al. 2019)
The ddPCR Workflow
Droplet Digital PCR is a 1-step process where reverse transcription and PCR are performed on individual droplets in the same reaction mixture. This process is considered advantageous because it can reduce inhibition of reverse transcription, in comparison to bulk methods.
7 Benefits of ddPCR Technology for Wastewater Surveillance

Faster Result
Assays using Droplet Digital PCR enable scientists to detect SARS-CoV-2 six days before clinical testing (Medema et al. 2020). 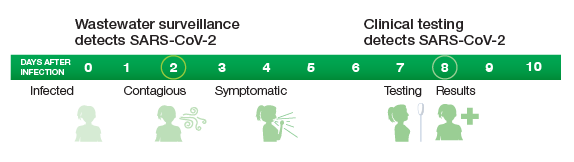

More Sensitive Results
Partitioning allows for enrichment of rare targets.
Detects 1 infected individual in 10,000
(Hart and Halden, 2020)

Absolute Quantification
Directly count DNA molecules and eliminate standard curves.

Inhibitor Tolerance
Endpoint PCR uncouples quantification from amplification and efficiency.

Variant Identification
Assays using Droplet Digital PCR can accurately discriminate and quantify multiple variants in a sample using a single-well test.

Cost Efficacy
Smaller sample requirements drive down cost.

Stability of Assay Design
Bio-Rad’s ddPCR Assays are designed to measure genes that are less mutated and more stable as amplification regions.

The PREvalence ddPCR SARS-CoV-2 Wastewater Quantification Kit measures the SARS-CoV-2 E and N2 genes and includes an assay for Murine Hepatitis Virus (MHV), a murine coronavirus. The test allows for all three targets to be detected in a single well.
Industries that May Benefit from ddPCR Surveillance
and Municipalities
Learn More about Droplet Digital PCR and Wastewater Surveillance
Watch the Webinar
Hear about scientists’ experiences building a successful wastewater surveillance system at Colorado State University.
Watch NowReferences
Gonzalez R et al. (2020). COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res 186, 116296.
Hart OE and Halden RU (2020). Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities and challenges. Sci Total Environ 730, 138875.
Jahne MA et al. (2019). Droplet digital PCR quantification of norovirus and adenovirus in decentralized wastewater and graywater collections: Implications for onsite reuse. Water Res 169, 115213.
Medema G et al. (2020). Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stages of the epidemic in the Netherlands. Environ Sci Technol Lett 7, 511–516.

