Immunotherapy has revolutionized cancer treatment, yet the drug discovery process to develop new biotherapeutic antibodies remains arduous. However, significant advances in antibody technology such as phage display have enabled fully human antibodies to be developed from animal-free libraries for just about any antigen.
The specificity of monoclonal antibodies (mAb) enables them to precisely target cancer cells by binding to cell surface markers. An example is rituximab, a chimeric mouse/human mAb that binds specifically to the cell surface B-cell protein CD20. Rituximab radically improves outcomes for B-cell malignancies like chronic lymphocytic leukemia. However, as rituximab is a chimeric antibody, the mouse portion can cause unwanted immunogenicity in human patients. While humanized antibodies have been able to reduce the issue of immunogenicity, fully human antibodies are more desirable for biotherapeutics as they further mitigate immunogenicity.
Generating Synthetic Libraries of Fully Human Antibodies in Fab Format
Phage display technology provides a method by which genes that encode proteins with specific characteristics can be identified and isolated from large DNA libraries in vitro. In the case of antibody phage display, antibody fragments featuring the antigen-binding site are displayed on the surface of the phage. These antibody fragments are typically heavy-chain variable domain (VHH), single-chain variable fragment (scFv), or antigen-binding fragment (Fab) format. The gene encoding the antibody fragment is contained within the phagemid vector of the phage on which the fragment is displayed, providing a physical link between genotype and phenotype. This collection of billions of phages, each displaying different antibody fragments on their surface, is referred to as a phage display library.
A technique called panning is used to screen a Fab phage display library for developable lead candidates (Figure 1).
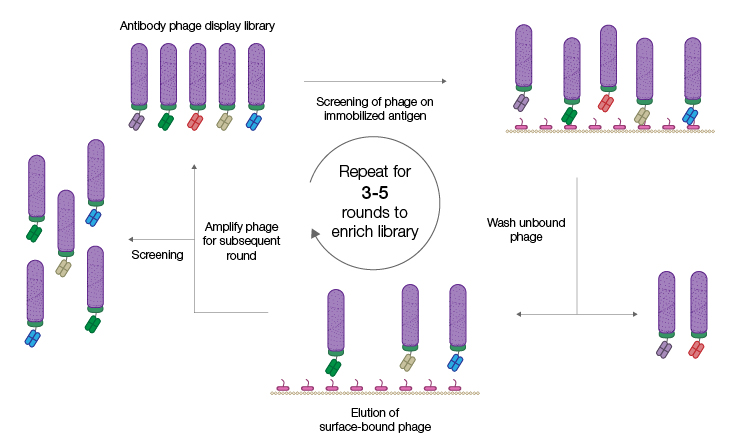
Fig. 1. Phage display selection by panning. The phage display antibody library is incubated with immobilized antigen. Phage displaying specific antibodies are in turn immobilized and phage displaying, nonspecific antibodies are removed by washing. Successive panning rounds result in an enriched population of phage displaying specific antibodies from which individual clones can be isolated. Figure recreated from Alfaleh et al. 2020 under a Creative Commons Attribution License (CC BY).
The immobilized antigen of interest is used as bait to capture phages displaying antibodies that specifically recognize that antigen. Nonbinders are removed by washing and phages that display antigen-specific antibodies are recovered and amplified by propagation in E. coli. Sequential rounds of panning result in a highly enriched population of phages that display antigen-specific antibody fragments. Individual Fab genes can be isolated from this enriched population, characterized, sequenced, and expressed as recombinant proteins. A major advantage of using an in vitro process like phage display is that it can be easily optimized to achieve the desired outcome. For example, other proteins can be spiked into the solution phase during panning to block out antibodies with undesirable cross-reactivity.
Transformative Technology
Synthetic antibody libraries can be designed to utilize only the human antibody germlines most suited to therapeutic antibody development. In addition, the amino acid distribution at each position in the complementarity-determining region (or hypervariable region) of the antibody can be controlled to omit sequence liabilities associated with developability issues or poor display rates in phage display.
The new Pioneer Antibody Library from Bio-Rad is, to our knowledge, the largest functional phage display Fab antibody library ever generated. The functional size as extrapolated by next-generation sequencing is believed to be 2.25 x 1011 antibodies (much bigger than the normal human immune repertoire). The correlation between library size and best-reported antibody affinities from published libraries (Hentrich et al. 2018) suggests that the larger the library, the more competition there is for binding to the target antigen and the higher the affinity of the antibodies that win that competition.
The vast size of the new Pioneer Library offers an even greater opportunity of identifying superior therapeutic antibodies with subnanomolar affinities to most antigens.
The Pioneer Library has been engineered with the proprietary SpyDisplay method (Figure 2), which harnesses SpyTag technology (also termed “molecular superglue”) to enable the rapid discovery of therapeutic antibody candidates. This novel method of displaying proteins eliminates the need for a subcloning step between selection and expression; after phage display library screening, the phagemid encoding the displayed protein is isolated from the selection strain and used to transfect a different E. coli expression strain (in the absence of helper phage), resulting in the expression of the displayed protein (Fab antibody) only. This makes the process less error-prone, faster, and more efficient, thereby accelerating the discovery workflow.
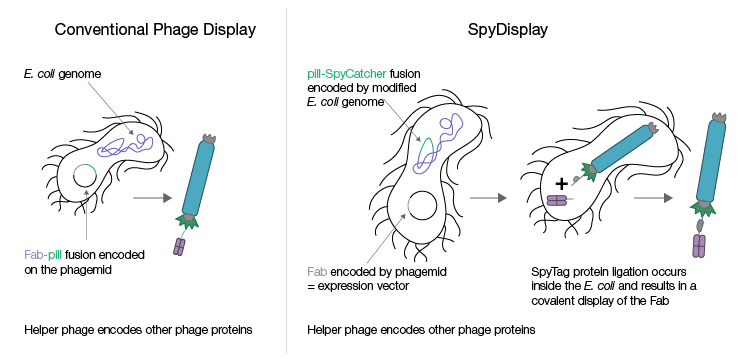
Fig. 2. Conventional phage display and SpyDisplay technologies. In conventional phage display, different vectors are used for selection during phage display and for the expression of individual clones after screening. Selection vectors typically encode the Fab antibody genetically fused to the pIII protein, and the expression vector encodes the Fab antibody alone. With SpyDisplay, one vector is used for both selection and expression. This vector encodes the Fab antibody with a SpyTag peptide fused at the C-terminus of the Fab heavy chain. A pIII-SpyCatcher fusion protein is encoded in the genome of the E. coli selection strain used for phage propagation. All other phage proteins are encoded by helper phage. Within the E. coli selection strain, the SpyTag-Fab fusion protein will spontaneously and covalently bind the pIII-SpyCatcher fusion protein during phage assembly.
Leading the Way
After phage display, the most suitable candidates from the Pioneer Library are then further screened to ensure that they are highly specific to the antigen of interest and to check for developability to ensure their suitability as drugs. For example, issues like low solubility and aggregation can cause production issues. Further selection then takes place and only the most suitable candidates identified from the initial hits are taken forward and optimized for drug development.
Find out how the Pioneer Antibody Library can support the discovery and development of therapeutic antibodies, and register your interest in this technology.
References
Alfaleh MA et al. (2020). Phage display derived monoclonal antibodies: from bench to bedside. Front Immunol 11, 1986.
Hentrich C et al. (2018). Monoclonal antibody generation by phage display: history, state-of-the-art, and future. In Handbook of Immunoassay Technologies. S. K. Vashist, J.H.T. Luong, ed. (Academic Press), pp. 47–80.

