Abstract
Surface plasmon resonance (SPR) is a well-established and important screening tool in the small molecule drug discovery workflow. Using the Bio-Rad ProteOn™ XPR36 protein interaction array system, an SPR biosensor platform, rapid optimization of immobilization conditions for a kinase target was carried out to ensure that the immobilized protein remains in an active state for the following screening step. Two kinase inhibitor compounds were screened against the kinase target to identify positive hits and to obtain binding kinetics. The whole experiment was completed in a fast workflow with a single sensor chip, showing the high performance of the ProteOn XPR36 system.
Introduction
Kinases are a family of proteins that catalyze the transfer of the gamma phosphate group of ATP or GTP to a substrate protein. As many as one third of all human proteins are thought to be substrates for protein kinases, making them one of the most important target groups for drug discovery.
Surface plasmon resonance (SPR) is useful to measure the affinities of potential kinase inhibitors. However, the kinetic rate constants can vary by several orders of magnitude even when compounds have similar affinities, and determination of affinities alone does not provide an adequate basis for lead selection.
It is well known that the conditions for immobilization on sensor chips are critical for maintaining the activity of protein kinases. Different methods for immobilizing kinases, such as premixing a kinase with an inhibitor and capturing a biotinylated kinase with a NeutrAvidin surface, have been reported to maintain the activity. However, when these specific conditions cannot be achieved, the general method of direct amine-coupling would be the choice for kinase immobilization. In the standard procedure of direct amine-coupling, the protein immobilization is carried out in low ionic strength buffer at a pH below the isoelectric point of the protein. This can be problematic for some kinases, as they are prone to losing their activity at low pH. Binding at higher pH to prevent protein inactivation can be achieved by increasing the concentration of protein injected. However, the ability to examine a range of different pH values to determine the right balance between the concentration of the protein at the surface and high enough pH to maintain the kinase activity is often a bottleneck for SPR analysis. Here we present a high throughput solution to quickly finding the conditions under which the kinase maintains activity.
Methods
Instrument and Reagents
Experiments were performed using the ProteOn XPR36 system with a GLH sensor chip. The ProteOn system features a 6 x 6 interaction array consisting of six vertical channels for ligand immobilization and six channels for analyte injection. Thus it is capable of simultaneously analyzing six ligand immobilization conditions together with six concentrations of analyte. Using the One-shot Kinetics™ approach, the ProteOn XPR36 system provides full kinetics for each of the immobilized ligand conditions. The experiment in this article utilized the One-shot Kinetics approach to achieve experimental optimization and interaction analysis with a single sensor chip.
Running buffer was 0.01 M HEPES pH 7.4, 0.15 M NaCl, 0.005% surfactant P20 with 5% DMSO, and 10 mM MgCl2. All experiments were performed at 25 ͦC. The kinase (MW 40 kD) and the three small molecule inhibitors (A 451 Da, B 413 Da, and C 420 Da) were all kindly supplied by Cancer Research UK.
Kinase Immobilization Conditions
The kinase was preincubated with ATP and then immobilized in all six vertical channels. Immobilization was performed at three different pH values, 4.5, 5.0, and 5.5 in 10 mM acetate buffer, each pH at two different protein concentrations.
Kinetic Binding Analysis
After deactivation, five different concentrations of each of the three compounds (500 nM to 6.2 nM) in threefold dilutions were injected simultaneously in the horizontal direction. Running buffer was injected in the sixth channel to enable real-time double referencing. Dissociation was monitored for 10 min. Regeneration of the ligand was not required between each compound due to fast dissociation rate of the compounds.
Data Analysis
The data were analyzed using ProteOn Manager™ 3.1 software. Binding curves were processed for baseline alignment, then interspot referencing and excluded volume correction were used. Each set of six reference-subtracted sensorgrams were globally fitted to curves described by the Langmuir 1:1 model.
Results and Discussion
Immobilization of the target kinase to the sensor chip surface was optimized by varying the pH and protein concentration during the immobilization step. Three acetate buffers with different pH values were tested at two different protein concentrations. To prevent ligand inactivation as a result of crosslinking at the binding site, the kinase was immobilized with 10 mM ATP for all six immobilization conditions. In this manner six different immobilization conditions (three different pH, each at two different protein concentrations) were tested simultaneously on one senor chip. Figure 1 shows the immobilization levels of the kinase at all six conditions.
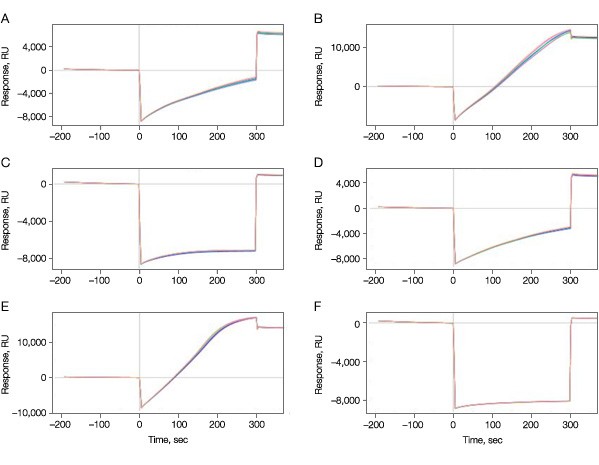
Fig. 1. Immobilization of the target kinase on the GLH sensor chip. A–C, 50 µg/ml; D–F, 100 µg/ml; A and D, pH 4.5; B and E, pH 5.0; C and F, pH 5.5.
As expected, the final immobilization levels were dependent on the pH values used, varying from as low as 479 RU at pH 5.5 to 14 kRU at pH 5.0 at 50 µg/ml protein.
The activity of the bound kinase was then determined for each immobilization condition by challenging it with small molecule A. The results in Figure 2 clearly show that the pH used during immobilization is critical with protein maintaining good activity at pH 5.0, but near 0 activity 0.5 pH units either side of this pH value.
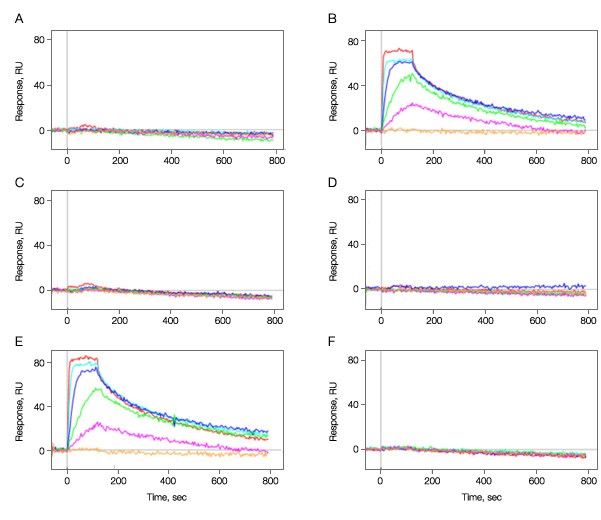
Fig. 2. Kinase immobilization at different pH conditions. Kinase was immobilized in all six vertical channels at different pH values. A–C, 50 µg/ml; D–F, 100 µg/ml; A and D, pH 4.5; B and E, pH 5.0; C and F, pH 5.5. Multiple concentrations of analyte A (threefold dilution series ranging from 500 nM to 6.2 nM) were injected in the horizontal channels at 100 µl/min for 2 min to test for binding activity. Dissociation was monitored for 10 min. The positive responses in graphs B and E show that the kinase immobilized at pH 5.0 was active in binding to compound A while that immobilization at a pH deviated by 0.5 units either side is not active. As graphs A–F correspond to the vertical channels L1–L6 in the ProteOn system, the channels L2 and L5 are the ones containing active kinase surfaces.
In the experiment, two compounds A and B were sequentially injected as analytes to interact with the active kinase surfaces in the vertical channels L2 and L5. The sensorgrams and fit curves from the Langmuir 1:1 model are shown in Figures 3A–3D. For compound A, the highest concentration 500 nM did not fit the Langmuir 1:1 model, possibly due to artifacts such as aggregate formation. Therefore it was excluded from the data fitting.
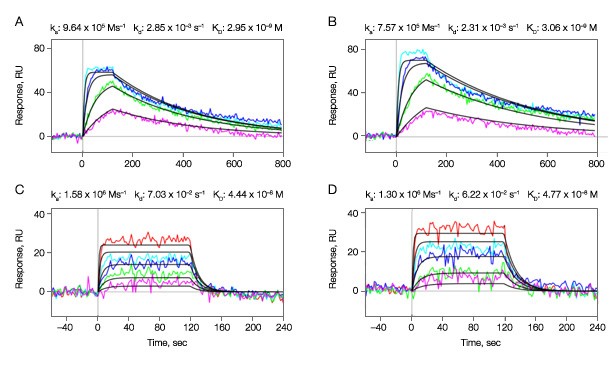
Fig. 3. Binding kinetics of compounds A and B to the active kinase surfaces in the vertical channels L2 and L5. Multiple concentrations of the analytes (compound A, threefold dilution series ranging from 167 nM to 6.2 nM; compound B, threefold dilution series ranging from 500 nM to 6.2 nM) were injected in the horizontal channels at 100 µl/min for 2 min to test for binding activity. A–B, compound A; C–D, compound B. A, C, sensorgrams obtained from the vertical channel L2; B, D, sensorgrams obtained from the vertical channel L5.
Kinetic Analysis
The kinetic analysis results for the two compounds are illustrated in Table 1. The data reveal that both small molecule inhibitors have similarly fast ka values, but show a greater range of dissociation rates that are two orders of magnitude different. The affinity constant KD in Table 1 is calculated from the kinetic constants (kd/ka).
Table 1. Kinetic constants for compounds A and B measured in two channels.
| Compound | Channel in ProteOn System | ka (Ms–1) | kd, (s–1) | KD (M) | Rmax (RU) | Chi2 (RU) |
| A | L2 | 9.64 x 105 | 2.85 x 10-3 | 2.95 x 10-9 | 60.6 | 13.7 |
| L5 | 7.57 x 105 | 2.31 x 10-3 | 3.06 x 10-9 | 70.9 | 20.2 | |
| B | L2 | 1.58 x 106 | 7.03 x 10-2 | 4.44 x 10-8 | 26.5 | 10.9 |
| L5 | 1.30 x 106 | 6.22 x 10-2 | 4.77 x 10-8 | 30.9 | 12.8 |
Equilibrium Analysis
It is evident from Figures 3C and 3D that injections for compound B reached the steady state. Thus equilibrium analysis is available to measure affinity constant KD directly. The results are shown in Figure 4 and Table 2.
Fig. 4. Equilibrium analysis for compound B.
Table 2. Affinity constant for compound B measured in two channels.
| Channel in ProteOn System | Rmax (RU) | KD (M) |
| L2 | 30.4 | 4.61 x 10-8 |
| L5 | 35.8 | 3.60 x 10-8 |
Summary
In this technical report we have demonstrated how the ProteOn XPR36 system can be used to identify the optimal conditions for kinase immobilization in a single injection cycle. By this means, accurate kinetic and affinity measurements for small molecules were successfully performed.
NeutrAvidin is a trademark of Thermo Fisher Scientific, Inc.



