Abstract
Protein purification often requires multiple iterations of individual column purifications, sample fractionation and visualization, and fraction pooling for the next column. Significant time is spent developing the purification strategy and using that information for subsequent protein purification cycles. A well-designed Multi-D method utilizes optimized individual column methods to create an automated and highly reproducible multistep procedure.
Here, we show the development of an automated Multi-D method on the NGC™ Chromatography System using a typical capture, intermediate, and polish purification workflow with an N-terminal small ubiquitin-like modifier-tagged C-terminal polyhistidine-tagged superfolder green fluorescent protein (SUMO-6xHis GFPsf), referred to as GFPsf hereafter. Our method development highlights the recovery and reproducibility of the automated purification process.
Figures
|
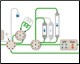
Figure 1. Schematic of the Multi-D setup used for the purification process |
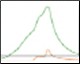
Optimization of IMAC Capture Step Figure 2. Pre-elution wash scouting |
|
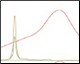
Optimization of Anion Exchange Intermediate Step Figure 3. pH and %B scouting |
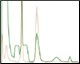
Development and Validation of Multi-D Method Figure 4. Single validation run of the Multi-D purification method |
|
Reproducibility of the Multi-D Method Figure 5. Three runs show the reproducibility of the Multi-D method |
Difference between Standard and Optimized Runs Figure 6. Elutions highlighting the importance of optimized individual purification steps |
|
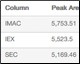
Percent Recovery of Purified Protein Table 1. High percentage of recovery between each column in the Multi-D method |
Introduction
Regardless of the application, scientists frequently perform multiple purifications of their protein of interest in order to generate sufficient quantities for their experiments. Depending on the purity needed, these workflows often require multiple iterations of single-column purifications, which require significant hands-on time for visualization, analysis, and fraction pooling. Multi-D chromatography represents a highvalue purification method due to its high reproducibility and “one-push” functionality.
Engineering a polyhistidine-tagged protein is such a commonly performed molecular biology technique that users would benefit from a quicker and more efficient purification process. Using Ni2+-immobilized metal affinity chromatography (IMAC) as the first capture step in a purification workflow can produce sufficiently purified material for certain applications; however, many workflows also incorporate intermediate and polishing steps to maximize the purity of the final product. GFPsf is a commonly purified recombinant engineered protein. Using the NGC Chromatography System with ChromLab™ Software, purification development of single-column steps can easily be optimized for purity and resolution. The information gained can be incorporated into a single Multi-D method by either editing one of the prewritten templates in the software or creating a custom method (see bulletin 6735 for more information).
Here, we illustrate the steps and reasoning behind the adaptation of a capture (Ni2+ IMAC), intermediate (anion exchange, AEX), and polish (size exclusion, SEC) workflow from individual column runs to a hands-free automated and reproducible Multi-D method. Typically, the eluate from the IMAC column is either diluted or dialyzed prior to being loaded onto the AEX column to lower the ionic strength to ~5 mS or less. In an automated purification, this is accomplished by using a desalting/buffer exchange column between the capture and intermediate steps. Therefore, the final Multi-D method incorporates IMAC, desalting, AEX, and SEC in a single automated workflow. We further demonstrate how the NGC Chromatography System and ChromLab Software can effectively be used to automate processes such as this.
Method and Results
System Configuration
Figure 1 depicts a three-tier NGC Quest™ 10 Plus Chromatography System configured with a sample pump, multiwavelength UV detector, two buffer inlet valves, two column switching valves (CSVs), and a single outlet valve. The recommended module placement and fluidics setup are depicted on page 6 of the NGC Chromatography Systems: Multidimensional (Multi-D) Chromatography Plumbing Guide (bulletin 6674). A multiwavelength UV detector was used to monitor GFP at both 495 nm and 280 nm during development and for recovery calculations. One of the CSVs was substituted with a buffer blending valve module for pH scouting during the AEX column optimization process but placed back into the system for the Multi-D method.
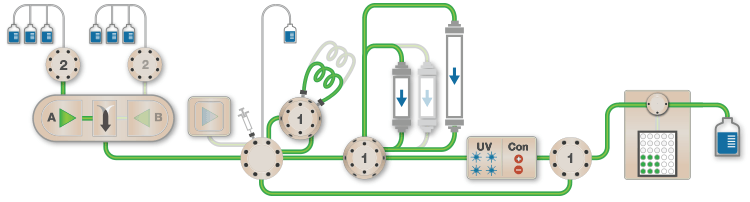
Fig. 1. Schematic of the Multi-D configuration of the NGC System. This schematic represents an NGC™ Quest 10 Plus System configured with a third tier, a sample pump, two buffer inlet valves, two CSVs, and an outlet valve. One of the CSVs is plumbed as a loop valve to capture the eluate from each column. Outlet valve port 2 is plumbed into the injection port of the inject valve to redirect eluate back into the system. All of the chromatography columns used were located on a single CSV. All of the static loops used in the Multi-D method were plumbed on the loop valve.
Fig. 2. Nuvia IMAC %B pre-elution wash scouting. Six scouting runs of the pre-elution wash from 0–25%B in 5%B increments. With increasing %B the leading shoulder of the 280 nm peak decreases without significant decreases in the GFPsf 495 nm trace up to 10%B. At 15%B we start to see significant GFPsf elution during the wash step and at 20%B all of the GFPsf has been eluted. A280 (—); A495 (—); %B (—).
Sample and System Preparation
Escherichia coli lysate containing GFPsf was clarified by centrifugation at 10,000 rpm for 10 min at 4°C and kept on ice. For the workflow optimization and Multi-D methods, buffer inlet valve A on the NGC System was primed with 1x PBS (IMAC buffer A, port 1), 0.025 M Tris, pH 8.0 (desalting and AEX buffer A, port 2), and 0.05 M Tris, pH 8.0 + 0.1 M NaCl (SEC buffer, port 3). Buffer inlet valve B was primed with 1x PBS + 0.5 M imidazole (IMAC buffer B, port 1) and 0.025 M Tris, pH 8.0 + 0.5 M NaCl (AEX buffer B, port 2). The sample pump was used to load sample during the single-step optimizations and for the IMAC purification during the Multi-D method. Prior to starting the method, the inject valve was placed into the inject position and the sample line was primed with the appropriate buffer A followed by a small volume of sample.
IMAC Capture Optimization
Utilizing the Affinity IMAC (Step Gradient) template in ChromLab Software, 2 ml of clarified lysate was loaded onto a 5 ml Bio-Scale™ Mini Nuvia™ IMAC Cartridge (Bio-Rad Laboratories, catalog #7800811) for each run. To better understand the binding and elution profile of GFPsf on the Nuvia IMAC Cartridge, the scouting feature within the software was used on the 3%B pre-elution column wash phase to generate a series of ten runs with increasing %B column wash steps (0–45% at 5% increments). Figure 2 shows a subset of those runs with an overlay of 0–25%B column wash where contaminating proteins are predominantly being eluted up to 10%B as seen by the decreasing front shoulder of the 280 nm elution peak. At 15%B, the GFPsf begins to elute from the column as seen by the decreasing intensity of the 495 nm peak. Based on these data, the 10%B pre-elution wash and an elution volume of 6 ml were determined to be optimal and used to generate the Multi-D method.
Desalting and AEX Intermediate Optimization
Using the Desalting template in ChromLab Software, a 50 ml Bio-Scale™ Mini Bio-Gel® P-6 Desalting Cartridge (Bio-Rad Laboratories, catalog #7325312) was used to remove IMAC buffer B with imidazole, placing the protein into a low ionic strength buffer for loading onto the AEX column. IMAC-purified protein (6 ml) was loaded onto the P-6 Desalting Cartridge and eluted by an isocratic flow of AEX buffer A to a final sample volume of ~14 ml.
The Anion Exchange template was used to load the desalted GFPsf (~14 ml) onto a 1 ml ENrich™ Q Column (Bio-Rad Laboratories, catalog #7800001). The optimal pH was determined using the scouting feature within ChromLab Software to determine the ideal binding strength and elution profiles (Figure 3A). As expected, we observed an increase in binding strength as the pH moved further away from the pI of the protein (pI = 6.2). The final %B concentration in the linear gradient step of the elution phase was scouted against, creating a series of runs from 20–70%B (Figure 3B). The scouting data show a shift in the elution peak during the 50%B endpoint that continued until the entire GFPsf peak was in the linear gradient during the 70%B linear gradient step in ~2 ml elution volume.
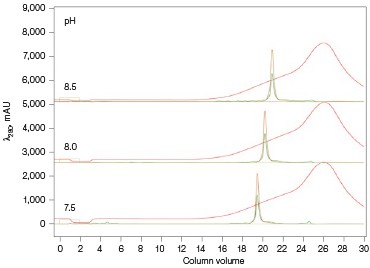
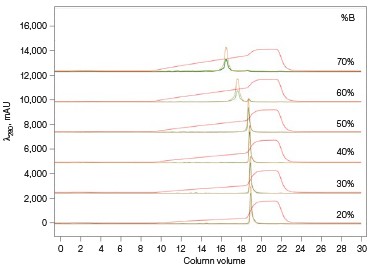
Fig. 3. ENrich Q AEX chromatography scouting. A buffer blending module was used to generate the pH scouting runs using Tris buffer for the ENrich Q Column scouting. A, pH scouting runs using Tris pH 7.5, 8.0, and 8.5. Tighter GFPsf binding is demonstrated by the increasing amounts of salt required to elute the protein as the pH increases further from the theoretical protein pI of 6.2. B, using pH 8.0 from the pH scouting, the linear gradient %B endpoint was scouted against starting at 20%B and increasing to 70%B in increments of 10%B. A GFPsf peak shift to the linear gradient begins at 50% and reaches completion by 70%B. A280 (—); A495 (—); conductivity (—).
SEC Polish Optimization
A 120 ml HiPrep 16/60 Sephacryl S-300 HR Column (GE Healthcare, catalog #17-1167-01) was used as the final column in the purification workflow. The purified protein was eluted with an isocratic flow of SEC buffer A (0.5 ml/min); no optimization was necessary.
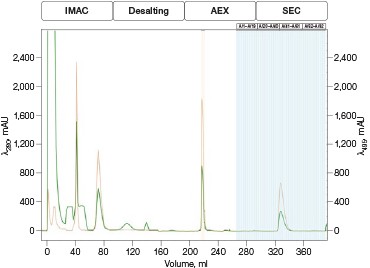
Fig. 4. Automated Multi-D IMAC purification method. Clarified GFPsf lysate (2 ml) was loaded onto a 5 ml Nuvia IMAC Cartridge using the sample pump. The column was washed with 10%B and the target protein was step-eluted and stored in a 10 ml static loop. This was loaded from the loop onto the 50 ml P-6 Desalting Cartridge, eluted isocratically, and collected into a 20 ml static loop. The 20 ml protein sample was loaded onto a 1 ml ENrich Q Column and eluted with a linear gradient. Using threshold collection, the eluate was shuttled into a 5 ml static loop, loaded onto a 120 ml HiPrep Sephacryl S-300 HR Column in a single injection, and eluted isocratically in 5 ml fractions. The blue lines represent final fraction collection of the purified protein product. A280 (—); A495 (—).
Multi-D Method Development and Validation
The phases in the 2-D Affinity (5 ml) – Desalting (50 ml) template were used as a starting point to create the fourcolumn Multi-D method. The majority of phases in the Multi-D method were copied over directly or replicated from the template method. Only the AEX elution phase was created by inserting steps into a standard phase (see bulletin 6735 for specific phase editing). Figure 4 shows a single validation run of the IMAC Multi-D purification workflow. Clarified lysate was loaded using the sample pump as previously described and fractions of purified GFPsf were collected only after the IMAC, desalting, AEX, and SEC separations were completed.
Discussion
For many scientists, the purification of protein samples for research purposes is necessary to obtain components essential to their downstream experiments. Frequently, it is a multiday labor-intensive process involving iterations of singlecolumn purifications, SDS-PAGE visualizations, and fraction pooling to advance the purification workflow. The majority of time spent on a purification workflow is in optimizing the binding and elution steps for each individual column. Once completed, the method can be followed as a simple routine for subsequent purification runs. As such, automating this process frees up a significant amount of time, which could be used more productively elsewhere. Equally important, small batch-to-batch variations between purification runs or by different individuals performing or monitoring the purification would be all but eliminated as the NGC Chromatography System reproducibly performs the same automated method over and over again (Figure 5).
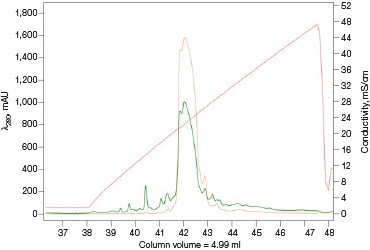
Fig. 5. Reproducibility of the Multi-D IMAC purification workflow method. Three automated IMAC runs were carried out to highlight the reproducibility of the purification strategy. Each method was completed in ~11.5 hours. A280 (—); A495 (—).
One of the most notable effects observed during the initial IMAC purification was the importance of the %B pre-elution column wash. Many scientists take for granted that weakly or nonspecifically bound proteins will wash off with just a small amount of imidazole in the wash. One benefit of using a GFP analog is the ability to track just the protein of interest at 495 nm using the multiwavelength detector module of the NGC System. This is an easier way to comparatively analyze runs without depending on SDS-PAGE visualizations and prevents interference with other possible co-eluting proteins and the imidazole in IMAC buffer B, which absorb at 280 nm. This allows for more accurate peak integration and recovery statistics.
Another important factor needed for any Multi-D method is the elution volume. A good starting point is to use a volume ~2x the column volume (that is, a 2 ml loop for a 1 ml column) because the volume of the column needs to be accounted for even in reverse column elution circumstances. The exception to this would be desalting columns. Assuming the entire eluted IMAC sample needs to be injected for desalting, a 50 ml P-6 Desalting Catridge would be required for the eluted IMAC protein stored in the 10 ml sample loop. Running the P-6 Cartridge with a 10 ml sample loop shows an elution volume of ~15 ml (data not shown). Since an AEX column follows the desalting, a larger volume presents no issues since the protein binds to the column and is concentrated in the process. A 20 ml loop would be sufficient to capture all of the protein from the P-6 Desalting Cartridge while maintaining the desalting effect.
Ion exchange offers several different variables to scout against but two of the most common are pH and %B endpoint of a linear gradient. Figure 3A shows a simple pH scouting using the buffer blending valve on the NGC System. Since the system automatically generates the desired pH required for each scouting run in addition to the linear gradient for elution using, in this example, Tris-HCl and Tris base, users do not need to make individual bottles of buffer A and B at each desired pH. As expected, the protein binds more tightly with increasing pH and increased displacement from the theoretical pI of 6.2. Peak integration shows a 2% difference in peak area for the three runs in both the 495 nm and 280 nm traces (data not shown) indicating no real binding or elution differences under the various pH conditions. For pH continuity with the PBS used for the IMAC purification, buffer at pH 8.0 was selected.
With this information, the elution gradient endpoint was scouted against. Eluting with a linear gradient typically gives the greatest chance of separation from contaminants. Templates frequently elute to 50% and then use a step gradient to 100% to strip any tightly bound proteins. Hence, it would be desirable to have the protein elute during the gradient portion of the elution and not the 100% wash at the end. Figure 3B shows the GFPsf elution peak starting to shift at the 50%B gradient endpoint, transitioning at 60%B with a small population of tighter binding GFP, and finally eluting by the 70%B linear gradient.
The final polishing column in the purification workflow is an SEC column. Optimization here is minimal since the sample volume helps dictate which SEC column will be used to ensure a single sample injection to maximize recovery. A sample volume of 5 ml requires the use of a 120 ml (16 x 600 mm) column. As it is the last column of the Multi-D method, concern about the elution volume is minimal because fractions are collected. Due to the low pressure limit and corresponding low flow rate, an alternative method to speed up the Multi-D method while maintaining recovery is using a smaller volume SEC column with a high pressure limit, such as an ENrich 70 (Bio-Rad Laboratories, catalog #7801070) or ENrich 650 Column (Bio-Rad Laboratories, catalog #7801650), and running multiple injections at a faster flow rate.
Once the individual steps of the purification workflow were optimized to provide the best peak separations, those conditions were combined with the information garnered about the elution volume to create the Multi-D method (see bulletin 6735 for the specific step editing of each phase).
Another consideration of Multi-D method generation is how each column will be eluted. Isocratic elutions make for easy loop collections by inserting change valve steps into a phase. Linear gradient elutions lend themselves more toward threshold elutions where the elution volume could be a little more variable due to the gradient endpoint and slope. Once the Multi-D method was created, the loop valve was equipped with a 10 ml loop on position 1 (IMAC elution), a 20 ml loop on position 2 (desalting elution), and a 5 ml loop on position 3 (AEX elution). The Multi-D method was validated to ensure the individual steps run correctly, the eluted samples are collected in the loops correctly, and the loop volume is sufficient. Multi-D method validation also helps verify that no further changes are required to the purification workflow. Figure 6 shows the difference between following a standard IMAC/Affinity template 3%B column wash and the optimized 10%B column wash. This helps underline the importance of optimizing each individual step of the purification when constructing a Multi-D method, as choices on the first column affect the efficiency and recovery of the final product. However, as Figure 4 demonstrates, once the method is validated, you can reproducibly generate purified protein sample. Table 1 demonstrates the high percentage of recovery between each column of the Multi-D method for the runs shown in Figure 5. This further validates the confidence in the reproducibility of the Multi-D method to produce the sample outcome every time in an automated, hands-free manner.

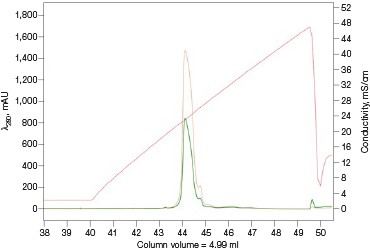
Fig. 6. Linear gradient elution of the ENrich Q AEX Column. Chromatograms of an automated IMAC purification emphasizing the ENrich Q Column linear gradient elution. A, pre-elution column wash phase during the IMAC purification at 3%B. The small shoulder peaks show co-eluting impurities that may risk a more contaminated sample. B, optimized 10%B pre-elution column wash phase during the IMAC purification step. The lack of small impurity peaks on the leading shoulder allows for a low threshold value to be used for collection of the eluate to the loop resulting in maximum recovery and purity. A280 (—); A495 (—); conductivity (—).
Table 1. Percent recovery statistics of the purified GFPsf protein.
| Column | Peak Area (495 nm) | % Recovery* |
| IMAC | 5,753.51 | 100.0 |
| IEX | 5,523.5 | 96.0 |
| SEC | 5,169.46 | 89.8 |
* Defined by the area of the peak of interest divided by the area of the IMAC peak, multiplied by 100.
The final automated method took ~11.5 hours to run from start to finish. This represents a significant amount of time savings compared to the traditional sequential approach, which would take 3–4 days per sample considering that, typically, SDS-PAGE is carried out and visualized between each step (in addition to potentially dialyzing the IMAC eluate overnight to remove buffer salts and imidazole). A significant amount of time in the Multi-D run is spent on equilibration and elution from the 120 ml HiPrep Sephacryl S-300 HR Column. Multiple injections on a smaller, more pressure-tolerant SEC column such as the ENrich 70 or ENrich 650 Columns could produce purified protein in a little over 4.5 hours, representing an even greater time savings. Another benefit of this automated workflow is the increased reproducibility between purification batches by removing the human error aspect of the purification. Ultimately, automated purification shows the most promise for consistent protein production at the laboratory scale.
Conclusions
- The Multi-D method provides hands-free functionality that enables the user to start a method and walk away
- Optimized methods for each step are vital to the construction of the Multi-D method to maximize reproducibility and recovery
- Multi-D methods can be constructed using the Tandem/2-D templates in ChromLab Software following the standard equilibration, sample application, column wash, and elution process of a single-column method
- Automated Multi-D methods enable users to consistently and reproducibly generate purified protein sample with confidence
The generation of an automated Multi-D purification workflow method is often seen as a barrier to achieving real automated protein purification. Several pieces of data are required prior to generating the Multi-D method. First, optimized conditions for each individual column in the purification must be established. These optimized conditions can be determined by using a variety of methods from simple univariate scouting, automated using the scouting feature within ChromLab Software, to more complex multivariate analysis utilizing DoE. Regardless of which method is used, the information about variables such as optimal pH, %B elution, sample loading, and flow rate must be incorporated into the Multi-D method. Observations of eluted sample volumes must be taken into account when programming elution phases, in addition to the size of the static loops needed for the capture steps between columns.
Once this information is known, the Multi-D method can be constructed either from scratch or by utilizing one of the prewritten 2-D templates in ChromLab Software, replicating its method phases with minor edits. Though the method creation in this study uses the example of an automated IMAC purification, most protein purification workflows can be converted into Multi-D methods.
Additional Resources
NGC Chromatography Systems: Multidimensional (Multi-D) Chromatography Plumbing Guide (bulletin 6674) — helps configure and plumb an NGC System for Multi-D applications.
Transforming a ChromLab Software 2-D Purification Template into an Automated Multidimensional (Multi-D) Purification Workflow (bulletin 6735) — helps adapt the preprogrammed method templates in ChromLab Software to customized Multi-D methods.
HiPrep and Sephacryl are trademarks of GE Healthcare.