Abstract
Droplet Digital PCR (ddPCR) enables easy multiplexing of numerous targets within each fluorescent channel. Multiplexing with Droplet Digital PCR allows for improved sensitivity and precision, as well as increased low-level detection in limited sample types like liquid biopsies. This is especially useful for increasing the sensitivity when detecting somatic mutations, edits, viruses, and other very rare targets. Multiple targets can be measured simultaneously by creating a unique endpoint fluorescence for each target. Amplitude-based multiplexing achieves this by varying primer and/or probe concentrations. In probe-mixing multiplexing, probes are mixed at desired concentrations to place targets in a defined position on a 2-D plot. This publication describes strategies for both approaches using the large portfolio of ddPCR Assays readily available through Bio‑Rad.
Figures
Introduction
There are many benefits to Droplet Digital PCR, including increased sensitivity, high-resolution copy number analysis, and accurate, precise molecule counting. Multiplexing is made simpler with Droplet Digital PCR due to its partitioning of samples into many thousands of droplet reactions for endpoint detection. Sample partitioning allows each droplet to be read and recorded individually so that each one has a unique fluorescent signature. This way, multiple fluorescent signatures can be read — all in one well. Often, when multiplexing primer and probe sets, assay efficiency imbalances during the PCR reaction can result in less-than-ideal performance (e.g., dropouts or missed targets in bulk reactions). However, partitioning with Droplet Digital PCR means each target is contained within a separate reaction vessel, reducing competition for resources. Finally, endpoint detection of each droplet post-PCR makes Droplet Digital PCR resistant to a wide range of efficiency differences between targets. As long as separation is achieved between droplet populations, an accurate count of the targets will be made.
There are two main types of multiplexing: amplitude multiplexing and probe-mixing multiplexing. For amplitude multiplexing, probe and/or primer concentrations can be varied to move the positive droplet clusters up or down on the axis, creating space for another assay’s results to be distinguished and clustered (Figure 1). For probe-mixing multiplexing, many unique fluorescent signatures are created by varying ratios of the probe in two different fluorescent configurations such as fluorescein (FAM) and hexachlorofluorescein (HEX) (Figures 2 and 3).
Expanding the number of targets multiplexed in a single well is further achieved by combining both discriminating and non-discriminating strategies. For example, each of the discriminated clusters can themselves have several possible targets within them that do not need to be discriminated. This mix of discriminating and non-discriminating multiplexing greatly increases the number of possible targets detectable in a single well.
Commercially available ddPCR multiplexing assays often use discriminating or non-discriminating strategies. Several products that employ non-discriminating multiplexing strategies are the ddPCR BRAF V600, ddPCR NRAS G12, ddPCR NRAS G12/G13, ddPCR NRAS Q61, and ddPCR KRAS Q61 Screening Kits (Bio‑Rad). Translocation multiplexing assays and a growing list of assays that use both discriminating and non-discriminating strategies are available as part of the Expert Design (EXD) Assays from Bio‑Rad.
Herein, we demonstrate three multiplexing examples that leverage each of the above strategies with the QX200 Droplet Digital PCR System.
Materials and Methods
Multiplexing was performed using TaqMan Assays with primers and probes labeled with FAM and/or HEX fluorophores and an Iowa Black FQ Quencher. Assays for amplitude multiplexing were ordered through Bio‑Rad Laboratories. Amplitude multiplexing was done using four unique copy number assays (two FAM and two HEX) in either 1x or 0.5x concentrations. Assays for radial multiplexing were purchased as custom assays. Probe mixes were created for the triplex as a 1:1 mix of the same primer and probe sequences in FAM and HEX in order to add a third cluster. The assay uses the following ratios of FAM to HEX: T1, 1:0; T2, 0:1; T3, 1:1. The 5-plex multiplex assays were also purchased as custom assays and used in the following ratios of FAM to HEX: T1, 1:0; T2, 3:1; T3, 1:1; T4: 1:3; and T5, 0:1.
The QX200 Droplet Digital PCR System was used with Droplet Generation Oil for Probes (catalog #1863005) and ddPCR Supermix for Probes (No dUTP) (#1863023). QuantaSoft Analysis Pro Software was used to analyze all data. Recommended thermal cycling conditions were used for copy number variation (CNV) assays (Figure 1 and Figure 2); recommended thermal cycling conditions for single nucleotide variation assays (SNV) or single nucleotide polymorphism assays (SNP) are shown in Figure 3.
Full assay concentration for the components of a 1x assay indicates 900 nM of each primer and 250 nM of each probe, unless otherwise stated. By adding a FAM assay to the reaction at the 1x concentration, adding the other FAM assay at 0.5x concentration, and doing the same with the two HEX assays, an upper and lower cluster can be created in the same well (four total targets per well) (Figure 1).
To move a cluster into a lower or higher position on the 2-D plot, it is not necessary for both the primer and probe concentration to be changed. Instead, similar results can be achieved by adjusting either the probe concentration or the primer concentrations. This is a simple, robust method for multiplexing four or more assays and can be used for copy number, gene expression, fusion gene detection, pathogen quantification, etc.
Different assays intrinsically vary in their ability to separate positive and negative clusters. This natural difference can be helpful when selecting an assay for each cluster position. In some cases, this difference may make adjusting primer and probe concentrations unnecessary. However, an iterative process may be needed in order to reach the desired cluster separation (e.g., using greater than 1x assay concentration in order to shift an upper cluster away from a lower one). It can be advantageous to increase the concentration of a higher amplitude assay if more than two assays will be used per channel (such as with a 6-plex multiplex ddPCR Assay). Increasing the concentration of the assay for the highest cluster beyond the 1x assay concentration would also be beneficial in cases where one of the assays does not perform well with 0.5x primer/probe concentration. Total amplification can be ensured by testing higher concentrations of high and low cluster assays alongside the 1x concentrations. Some optimization and verification of performance may be required if decreasing the assay concentration below the recommended amount.
Both amplitude and probe-mixing multiplexing are simple methods for quantifying multiple targets within a single well. One benefit of using probe-mixing multiplexing over amplitude multiplexing is that it allows the classification of low- or mid-amplitude droplets, termed “rain,” that may occur between the negative cluster and the positive cluster. This feature is ideal for rare mutation detection, low-concentration samples, and other difficult sample types.
Results
Amplitude Multiplex Droplet Digital PCR
The 2-D plots generated after performing each multiplex ddPCR experiment show each distinguished target/target set within unique clusters. In Figure 1, data from the multiplex amplification of four CNV assays targeting genes in human DNA are shown. The positions of the various clusters were altered by varying primer and probe concentrations. In Figure 1 the concentrations of both the primer and probe were decreased by half using four CNV assays (two FAM assays and two HEX assays).
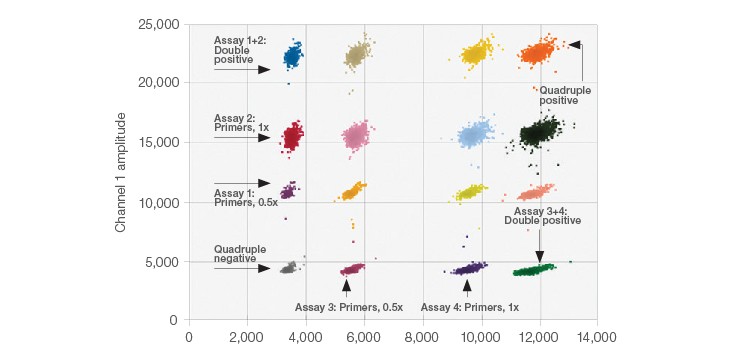
Fig. 1. 2-D Amplitude multiplexing plot. The expected concentrations and clear separations were achieved for all 16 clusters.
Probe-Mixing Multiplexing Droplet Digital PCR
The probe-mixing multiplexing strategy is an easy way to incorporate up to six clusters on a 2-D plot. In Figure 2, an example of this strategy is shown using a mixture of fluorophores for one of the target sequences. This creates a third cluster (T3) located in the center of the 2-D plot between the two single channel positives (T1 and T2) (i.e., a triplex). This triplex uses 50% FAM and 50% HEX, totaling the 250 nM final probe concentration. The other two targets are at 1x concentrations and use either 100% FAM or 100% HEX. The probe mix triplex setting in the QuantaSoft Analysis Pro Software was used to classify and analyze the target clusters.
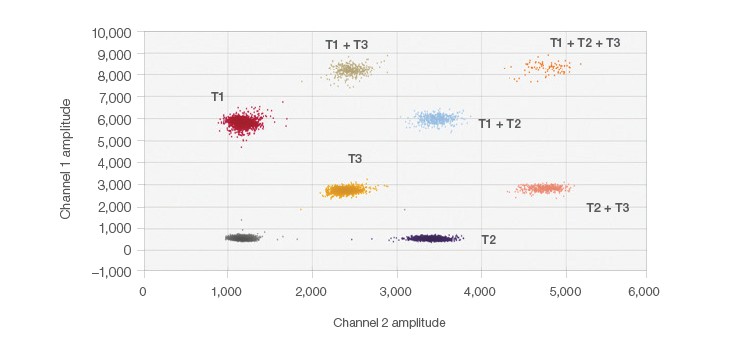
Fig. 2. Probe-mixing multiplex 2-D plot. Clearly distinguishable clusters are shown here in this example of probe-mixing multiplexing of three targets in one well.
Figure 3, instead, shows the results from the 5-plex assay using the probe-mixing multiplex strategy to create target clusters based on the ratios of FAM and HEX fluorescent probes for SNPs that exist in the genome. The probe mixtures create several clusters, arranged radially on the 2-D plot (Figure 3). This was achieved by adding mixtures of probes containing the same nucleotide sequence, but different fluorophores (FAM and HEX). By varying the fluorophore ratios for each of the probes, each of the five targets exists in a unique location on the plot, yet no overlap occurs. QuantaSoft Analysis Pro Advanced Classification Mode was used to analyze this multiplex ddPCR experiment. This multiplexing strategy is ideal for differentiating somatic mutations or pathogens since positive samples would only be positive for one or two of the targets, simplifying thresholding and analysis.
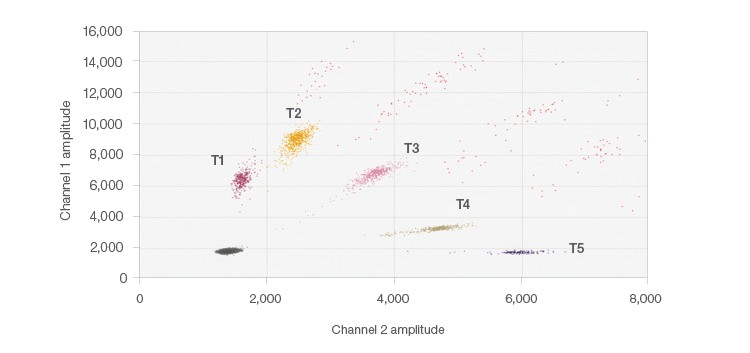
Fig. 3. 5-plex probe-mixing multiplex 2-D plot. The single positive cluster is labeled for each target; multiple occupancy droplets appear in orange. Each of the five targets was designed to detect one of two potential SNPs at each location. A similar strategy can be employed for mutation detection.
Conclusion
Multiplexing enables more sensitive detection of many targets from a small sample, saves money, and increases sample throughput. Combined with Droplet Digital PCR, multiplexing is effective and can be achieved by applying the two strategies demonstrated above. We have been able to show the ease of multiplexing three, four, and even five targets — with only two fluorescent channels. Further, QuantaSoft Analysis Pro Software offers straightforward analysis for both of the simple multiplexing strategies described here. Using these two strategies makes developing multiplex workflows simple and robust. A variety of multiplexing strategies continue to be designed in order to fit other assay types and unique needs (e.g., drop-off probes, EvaGreen®, or mixing EvaGreen® and probe assays together).
For more details on multiplexing data analysis, see the QuantaSoft Analysis Pro Software Instruction Manual (bulletin 6827).
For more information about assay design, see our Droplet Digital PCR Applications Guide (bulletin 6407).
The QX200 Droplet Digital PCR System, and the consumables and reagents designed to work with this system, and/or their use is covered by claims of U.S. patents and/or pending U.S. and non-U.S. patent applications owned by or under license to Bio‑Rad Laboratories, Inc. See bio‑rad.com/en-us/trademarks for details. Purchase of the product includes a limited, non-transferable right under such intellectual property for use of the product for internal research purposes in the field of digital PCR only. No rights are granted for diagnostic uses. No rights are granted for use of the product for commercial applications of any kind, including but not limited to manufacturing, quality control, or commercial services, such as contract services or fee for services. Information concerning a license for such uses can be obtained from Bio‑Rad Laboratories. It is the responsibility of the purchaser/end user to acquire any additional intellectual property rights that may be required.
EvaGreen is a trademark of Biotium, Inc. Bio‑Rad Laboratories, Inc. is licensed by Biotium, Inc. to sell reagents containing EvaGreen Dye for use in real-time PCR, for research purposes only.




