Within one year, Ebola virus disease (EVD) has gone from relative anonymity to a worldwide source of fear. This disease has been brought to the forefront of the public’s attention due to its gruesome symptoms (which include vomiting, diarrhea, and hemorrhagic bleeding), high mortality rate (up to 90% according to some reports), and lack of specific treatment options. On March 23, 2014, the World Health Organization (WHO) was first notified of an outbreak in Guinea, and the virus has since then severely affected the West African countries of Guinea, Sierra Leone, and Liberia. The spread of EVD to multiple countries, including the U.S., has led the WHO to identify EVD as a “public health emergency of international concern.” Although over 5000 EVD-related deaths have been reported worldwide as of November 2014, the WHO states this number is likely underestimated.
Ebola’s attack on the immune system
Four different viruses of the genus Ebolavirus can cause EVD. Of these, the Ebola virus (EBOV, formerly known as the Zaire Ebola virus) is the most severe and has caused the greatest number of outbreaks over the past 40 years. The key contributor to the high mortality rate is the ability of EBOV to specifically reduce immune system function, which allows the virus to rapidly infect cells and tissues throughout the body.
EBOV consists of a single-stranded negative-sense 19 kb RNA genome that contains seven genes (Figure 1). As with many viruses, the virions attach to surface receptors on the cell membrane. The virion is then internalized via receptor-mediated endocytosis (White and Schornberg 2012), though there is some debate over the exact mode of entry into the host cell. The viral membrane fuses to the endosomal membrane and the viral contents are released into the cytosol. The RNA polymerase transcribes mRNA molecules, which are translated into the structural and nonstructural proteins of the virus. The structural proteins, including VP24, VP30, VP35, and VP40, and the RNA genome accumulate near the cell membrane. When instructed by the viral nucleoprotein, the virions bud off from the cell and infect neighboring cells, repeating the above process (Figure 2). The structural glycoproteins (GPs) on the surface of the virus help it bind to and infect target cells and are an important treatment target in recent experimental antibody-based treatments and vaccines (as detailed further in the Experimental treatment of EBOV with monoclonal antibodies and Ebola vaccines — current clinical trials and improved production methods sections, respectively).
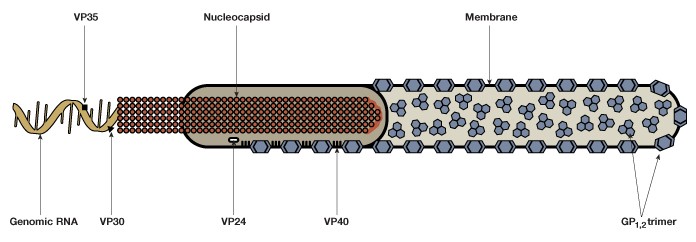
Fig. 1. The Ebola virus is composed of a single-stranded negative-sense genomic RNA, a nucleocapsid, VP structural proteins, glycoprotein complexes (GP1,2), and an outer membrane.
EBOV replication is particularly efficient in immune cells (including macrophages, monocytes, and dendritic cells). The immune cells carry the virus to lymph nodes, where the virus replicates further and spreads through the bloodstream and lymphatic system. As the EBOV spreads throughout the body, it infects the endothelial cells that line the blood vessels and secretes small soluble GPs, which form trimeric complexes to attach the virus to the endothelial cells. The GPs also inhibit neutrophil activation by forming a dimeric protein that interferes with neutrophil signaling. The damage to these cells leads to an excessive production of cytokines and reduces vascular integrity, which can lead to internal and external bleeding along with decreased blood clotting.
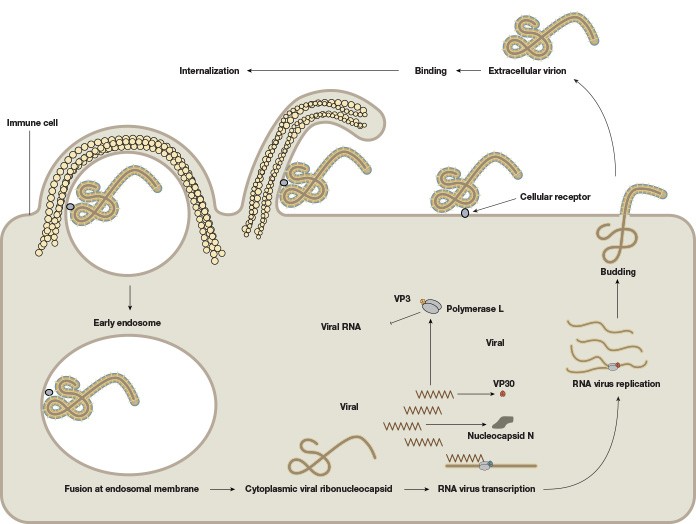
Fig. 2. The life cycle of the Ebola virus. The viral membrane fuses to the endosomal membrane, and the viral contents are released into the cytosol. The cytoplasmic viral nucleocapsid undergoes mRNA transcription, followed by translation of the mRNA into viral proteins. These viral proteins accumulate near the membrane in viral particles (virions), which bud off from the cell. The extracellular virion binds to and fuses with a neighboring cell, repeating the process.
Infected macrophages and lymphocytes undergo programmed cell death, which further weakens the immune system. Furthermore, the VP24 and VP35 structural proteins of EBOV interfere with the innate immune system by inhibiting the cell’s production of and response to interferons, proteins that have antiviral properties, which allows EBOV to spread throughout the body uninhibited by the immune system. Death, if it occurs, typically takes place within 6-16 days after the onset of symptoms.
Diagnosis of EBOV — improving access to real-time PCR in West Africa
Real-time PCR, which detects the presence of the EBOV genetic material, is the most specific and practical method for diagnosing EVD in the early stages. However, until recently, many treatment units did not have quick access to PCR-based laboratories. Health workers would have to send blood samples to outside clinics and wait 2-5 days for results. This delay can compromise early care and isolation of infected individuals, which is especially problematic considering EVD often causes death within two weeks of symptom onset.
Therefore, the WHO has been focused on increasing the number of PCR-based labs in the hard-hit capital cities of Liberia and Sierra Leone (Monrovia and Freetown, respectively). In October 2014, the U.S. Navy opened an Ebola mobile laboratory near the Island Clinic, a treatment unit in Monrovia that can detect EBOV in blood samples within 3 hours using PCR. According to Dr. Pranav Shetty, emergency medical coordinator for the non-profit International Medical Corps (IMC), up to 80 samples can be screened per day, and over 500 samples (about half of which were positive for EBOV) were tested at this unit within the first month of operation.to 80 samples can be screened per day, and over 500 samples (about half of which were positive for EBOV) were tested at this unit within the first month of operation.
Before isolating the RNA from the blood, the researchers use processes such as gamma irradiation to eliminate the infectivity of the blood sample. The RNA is then isolated and real-time PCR is used to confirm the presence of the virus. Thus, individuals who have the virus are identified quickly and accurately, which helps improve treatment of infected individuals and minimize their exposure to others.
Paper-based antibody detection — rapid diagnosis without electricity
Although antibody detection (often using antigen-capture enzyme-linked immunosorbent assay [ELISA] in the laboratory setting) is not as sensitive as real-time PCR, new advances in antibody screening may aid diagnosis in regions that do not have access to PCR testing. In cooperation with Corgenix, researchers at Tulane University in New Orleans, LA, are experimenting with a rapid diagnostic test strip containing dye-tagged antibodies that bind to a specific EBOV protein. Unlike real-time PCR, which requires a needle-drawn blood sample, this test strip will only require a drop of blood applied directly from a pricked finger. Furthermore, the test strip does not require the steady supply of electricity necessary to run PCR machines, making it a more convenient option for remote areas. Senova, a company based in Weimar, Germany, has also developed a similar test and is running samples in parallel with the standard PCR procedure to determine their test’s accuracy.
However, Robert Garry, a virologist who is coordinating the project at Tulane, indicated that even if the test is not as sensitive as PCR, it will still be helpful. According to Garry, the tests could be used as a screening tool in hard-to-reach villages to determine whether further intervention is needed to contain the virus.
“If you test 10 people and none show up positive, you can move onto the next village,” said Garry.
Another group of scientists at the Wyss Institute for Biologically Inspired Engineering in Boston, MA, have developed a way of detecting strains of the Ebola virus using pocket-sized blotting paper. This technology involves embedding freeze-dried cell-free synthetic gene networks, along with the dNTPs, tRNAs, amino acids, and buffers needed for gene transcription and translation, onto a paper platform. To detect the presence of EBOV in a saliva or blood sample, an RNA “trigger molecule” is programmed so that it reacts (changes the color of the paper) if the EBOV is present in the sample and embedded into the paper. The test takes 30 minutes to produce a result, costs only $20, is easily transported, and is stable at room temperature for up to one year, making the test a potentially convenient option for distribution in remote areas that do not have reliable access to electricity.
Keith Pardee, the lead scientist investigating this technology, states that although these tests have not been validated in Ebola patients, the proof-of-principle demonstrations in the laboratory and the “number of technologies under development,” will help improve the sensitivity of this test for future field application in detecting Ebola and potentially other diseases.
Experimental treatment of EBOV with monoclonal antibodies
Current treatment of EVD primarily involves rehydration and symptom management. Of the experimental treatments that specifically target the genome or proteins of EBOV, monoclonal antibody (mAb) therapy is the only one that has been successful when administered more than 24 hours after exposure in non-human primates. In 2013, two groups of researchers showed that approximately 50% of non-human primates infected with EBOV survived after administration of MB-003 or ZMAb, whereas their untreated controls died of the disease. MB-003 and ZMAb are each composed of three different EBOV glycoprotein-specific antibodies that bind to and inactivate a unique glycoprotein epitope in the EBOV, thereby preventing the virus from infecting healthy cells (Qiu et al. 2014).
Although the studies investigating the MB-003 and ZMAb antibody cocktails were promising, scientists from Mapp Biopharmaceutical sought to improve on these results. In collaboration with Kentucky BioProcessing, the Public Health Agency of Canada, and U.S. National Institute of Allergy and Infectious Diseases, the researchers investigated several combinations of mAbs to determine which resulted in the best survival rates in EBOV-infected rhesus macaques. The researchers found that ZMapp, a trio of chimerized monoclonal antibodies (c2G4 and c4G7 from ZMAb and c13C6 from MB-003) was effective in treating 18 rhesus macaque primates infected with the Kikwit Congolese variant of EBOV, whereas the three untreated control primates died from the EBOV. Immunoglobulin G (IgG) ELISA assays using two variants of gamma-irradiated EBOV (the Kikwit Congolese and Guinean variants) as the capture antigen showed that ZMapp also binds to the Guinean strain of EBOV in cell culture, suggesting this drug may be effective across multiple EBOV strains and potentially respond to escape mutations (mutations in the genome that allow the virus to escape the immune system).
Making ZMapp available to the public with magnifection
Limited doses of the drug have been used experimentally to treat individuals infected with EBOV. As of October 2014, five patients who received the treatment have survived, and two have died. Although the supplies of ZMapp have been exhausted, the U.S. government has signed a multimillion dollar contract with Mapp Biopharmaceutical to produce enough ZMapp for early stage clinical safety studies. Mapp will also collaborate with the US Biomedical Advanced Research and Development Authority (BARDA) to increase efficiency of its manufacturing process to improve production yield.
Until recently, developing such a large quantity of mAbs using processes such as stable genetic transformation and transient infection with viral vectors would have been time-consuming and prohibitively expensive. However, a process called magnifection is capable of synthesizing this product in the Nicotiana benthamiana tobacco plant using Agrobacterium as an infective agent. Agrobacterium delivers viral replicons to different parts of the plant, producing a high-yield mAb product in approximately one month (as opposed to 1-2 years with the previously mentioned methods). The process can also accommodate high-level co-expression of multiple and large proteins, making the process optimal for production of a chimerized multiple-antibody protein such as ZMapp.
For magnifection of ZMapp, researchers inserted the genes that code for the three mAbs into viral vectors. These vectors were loaded into Agrobacterium cultures, which were used to infect the N. benthamiana plants (Figure 3). After growing the plants for 4 weeks in an enclosed growth culture room, the researchers mixed overnight-grown Agrobacterium cultures into infiltration buffer, dipped the aerial parts of the entire plant into this bacterial/buffer solution, and applied a vacuum. After infiltration, plants were grown for another 8 days, at which point the leaf tissue was extracted in a juicer and the antibodies were purified. This expression of human antibody proteins in plants allow for rapid, high-yield production of full-size monoclonal antibody, which not only allowed the scientists from Mapp Biopharmaceutical to experiment with several combinations of mAbs, but may also allow wide availability of ZMapp at a reasonable cost for treatment of EVD in the future.
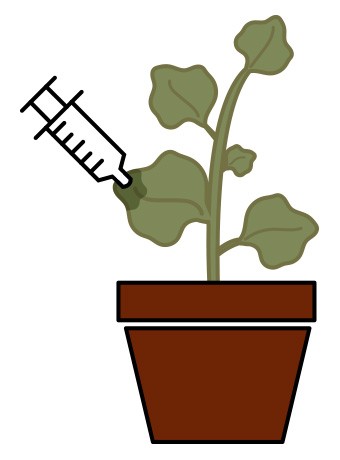
Fig. 3. ZMapp, a promising new experimental treatment for Ebola, is synthesized through magnifection of the Nicotiana benthamiana tobacco plant using Agrobacterium as an infective agent.
Ebola vaccines — current clinical trials and improved production methods
Several vaccine candidates, including replication-deficient adenovirus vectors, replication-competent vesicular stomatitis virus (VSV)-based vaccines, virus-like particle vaccines, and recombinant human parainfluenza virus 3-based vaccines, have been developed and tested in primates prior to the 2014 outbreak. However, the current situation in West Africa has hastened the efforts of U.S. government officials to fast-track production of vaccine candidates. Because conventional trials (in which subjects are given the vaccine and then exposed to the virus) in humans are not ethical or feasible, the FDA has established the “animal rule,” which would allow vaccine approval based on an animal model combined with evidence of safety and a potent immune response in humans. Respective clinical trials for the replication-deficient cAd3-EBO Z vaccine and the replication-competent VSV ZEBOV vaccine, both of which target the surface glycoproteins of the EBOV, began in October 2014.
If effective, the vaccines may be given to individuals at high risk of contracting the virus, such as healthcare workers. However, EBOV is extremely virulent, which may make development of a universally effective vaccine difficult. According to John Dye, chief of the viral immunology branch at the U.S. Army Medical Research Institute of Infectious Diseases, EBOV is “a survivor.”
“It does what it can to avoid the human immune system,” said Dye.
Several novel Bio-Rad products may help increase efficiency of future vaccine production, perhaps improving the ability to modify vaccines for viral strains with new escape mutations. The QX200™ Droplet Digital™ PCR (ddPCR™) System quantifies DNA and RNA without relying on rate-based measurements or calibration curves and can detect the presence of a single target molecule even among a large number of microreactions. Recent research has shown that using ddPCR increased single-stranded and self-complementary adeno-associated viral vector genome titers up to 5-fold versus conventional qPCR (Lock et al. 2014). Furthermore, the intra- and interassay precision was superior for ddPCR-based genome titering than for qPCR, making this technology an attractive option for accurate monitoring of early-stage production of the viral vectors for potential Ebola vaccines and for analyzing the viral vector genome in biological tissues.
Future Directions
The 2014 outbreak of EVD is the largest documented and has led to tens of thousands of illnesses worldwide. Rapid diagnostic capacity and effective pharmaceutical agents that specifically target the virus and can respond to escape mutations will be important for managing current and future outbreaks. For example, Bio-Rad’s Bio-Plex® Multiplex System uses bead-based technology to simultaneously multiplex up to 500 different assays and 2160 samples in 3 hours. This thorough diagnostic analysis may make the device particularly useful for characterizing the serological profile of individuals affected by new viral strains. Indeed, the Bio-Plex System was used to measure serological and cytokine profiles in individuals infected with Bundibugyo ebolavirus, a new species of the Ebolavirus discovered in a 2007 outbreak in Uganda (Gupta et al. 2012). In this study, scientists measured serum levels of 17 cytokines and chemokines using a single customized kit. They found that the serum circulating cytokine profiles in individuals infected with this species were significantly different from individuals infected with Zaire and Sudan ebolaviruses. Such precise diagnostic analysis will better enable scientists to identify and develop more specific therapies and vaccines for future infections of all types.
References
Gupta et al. (2012). Serology and cytokine profiles in patients infected with the newly discovered Bundibugyo ebolavirus. Virology 423, 119–124.
Lock M et al. (2014). Absolute determination of single-stranded and self-complementary adeno-associated viral vector genome titers by droplet digital PCR. Hum Gene Ther Methods 25(2),115-25.
Qiu X et al. (2014). Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514(7520),47-53.
White JM and Schornberg KL (2012). A new player in the puzzle of filovirus entry. Nature Rev Microbiol 10, 317–322.
The QX200 and/or its use is covered by claims of U.S. patents, and/or pending U.S. and non-U.S. patent applications owned by or under license to Bio-Rad Laboratories, Inc. Purchase of the product includes a limited, non-transferable right under such intellectual property for use of the product for internal research purposes only. No rights are granted for diagnostic uses. No rights are granted for use of the product for commercial applications of any kind, including but not limited to manufacturing, quality control, or commercial services, such as contract services or fee for services. Information concerning a license for such uses can be obtained from Bio-Rad Laboratories. It is the responsibility of the purchaser/end user to acquire any additional intellectual property rights that may be required.
The Bio-Plex suspension array system includes fluorescently labeled microspheres and instrumentation licensed to Bio-Rad Laboratories, Inc. by the Luminex Corporation.

