RNA sequencing (RNA-Seq) is a fundamental technique for transcriptome profiling, facilitating the detection of gene expression changes across sample groups. Traditional RNA-Seq of complex tissues, however, is not capable of deciphering the unique biological processes that take place within each cell. As such, single-cell RNA-Seq (scRNA-Seq) has emerged as a compelling alternative for the comprehensive analysis of RNA molecules from individual cells, uncovering detailed biological insights from heterogeneous cell populations, which are unobtainable through bulk RNA-Seq analysis.
Offering a high-throughput and multidimensional analysis of individual cells, single-cell RNA-Seq analysis provides a high-resolution view of gene expression variability which, in turn, allows the characterization of cell subpopulation classification and cell heterogeneity. This approach has been used extensively in several cancer research applications including the identification of rare cell types from otherwise homogeneous cell populations, such as malignant tumor cells within a tumor mass, monitoring tumor cell evolution and drug resistance (Haque et al. 2017).
Additional scRNA-Seq applications include immune cell profiling and the characterization of nerve cell expression and function, providing insights into complex neurological disorders (Awuah et al. 2023). Moreover, during the recent COVID-19 pandemic, single-cell approaches were used to identify immune correlates of disease severity in human tissue, where comparison of bronchoalveolar lavages of patients infected with COVID-19 generated immune profiles associated with disease status (Van de Sande et al. 2023). scRNA-Seq can also be combined with genomic and proteomic data to describe more complex cellular heterogeneity than transcriptome data alone (Choi et al. 2020, Lim et al. 2024).
As part of single-cell transcriptomics, 3 RNA-Seq has emerged as an advanced technique for gene expression analysis, offering several advantages over full-length RNA-Seq. Unlike standard RNA-Seq, mRNA sequences for 3′ RNA-Seq are not fragmented prior to reverse transcription; instead, cDNAs are synthesized only from the 3′ end of the mRNAs, producing a single cDNA copy for each transcript. This approach ensures that the number of sequencing reads corresponds directly to the number of transcripts of a specific gene, providing unbiased coverage for both longer and shorter transcripts (Ma et al. 2019).
Commonly Used 3′ RNA-Seq Methods for Single-Cell Analysis
There are several 3′ RNA-Seq methods currently available, varying in ease of workflow, performance, and cost.
One commercially available technology utilizes a droplet-based method to generate gel bead-in-emulsion partitions that encapsulate individual microreactions. These individual samples are barcoded to separately index each cell’s transcriptome. This approach offers high-quality data but is dependent upon expensive instrumentation, which can be prohibitive for researchers entering the single-cell sequencing field.
Instrument-free technologies offer comparable performance and scalability without the need for investing in specialized instrumentation. Examples include plate-based combinatorial barcoding and vortex emulsion methods of droplet generation. However, there are still concerns regarding workflow complexity and, in some cases, data quality.
ddSEQ – A Cost-Effective Alternative Solution for High-Quality Single-Cell Transcriptome Profiling
Bio-Rad has recently leveraged their proven ddSEQ droplet technology to develop the ddSEQ 3′ Single-Cell RNA-Seq Kit, which enables streamlined high-quality whole transcriptome analysis (WTA) of 500–5,000 individual cells with high sensitivity and cell capture. Designed for use with the ddSEQ Isolator for Single-Cell Sequencing, cells are encapsulated in a rapid droplet partitioning step, followed by an efficient assay workflow (Figure 1). This droplet-based microfluidic approach reduces the volume of sample required, thereby minimizing associated costs and sample preparation workflows.
The resulting quality control and data analysis is enabled by the accompanying Omnition Analysis Software, version 1.1, which is a flexible and robust open-source pipeline analysis tool. This bioinformatics pipeline software allows batch analysis of samples, saving time and improving process efficiency.
A key benefit of the ddSEQ Single-Cell 3′ RNA-Seq Kit is its low cost, which allows researchers to stretch their budgets while delivering high quality scRNA-Seq data. The simple workflow reduces processing time, providing increased flexibility to users’ schedules.
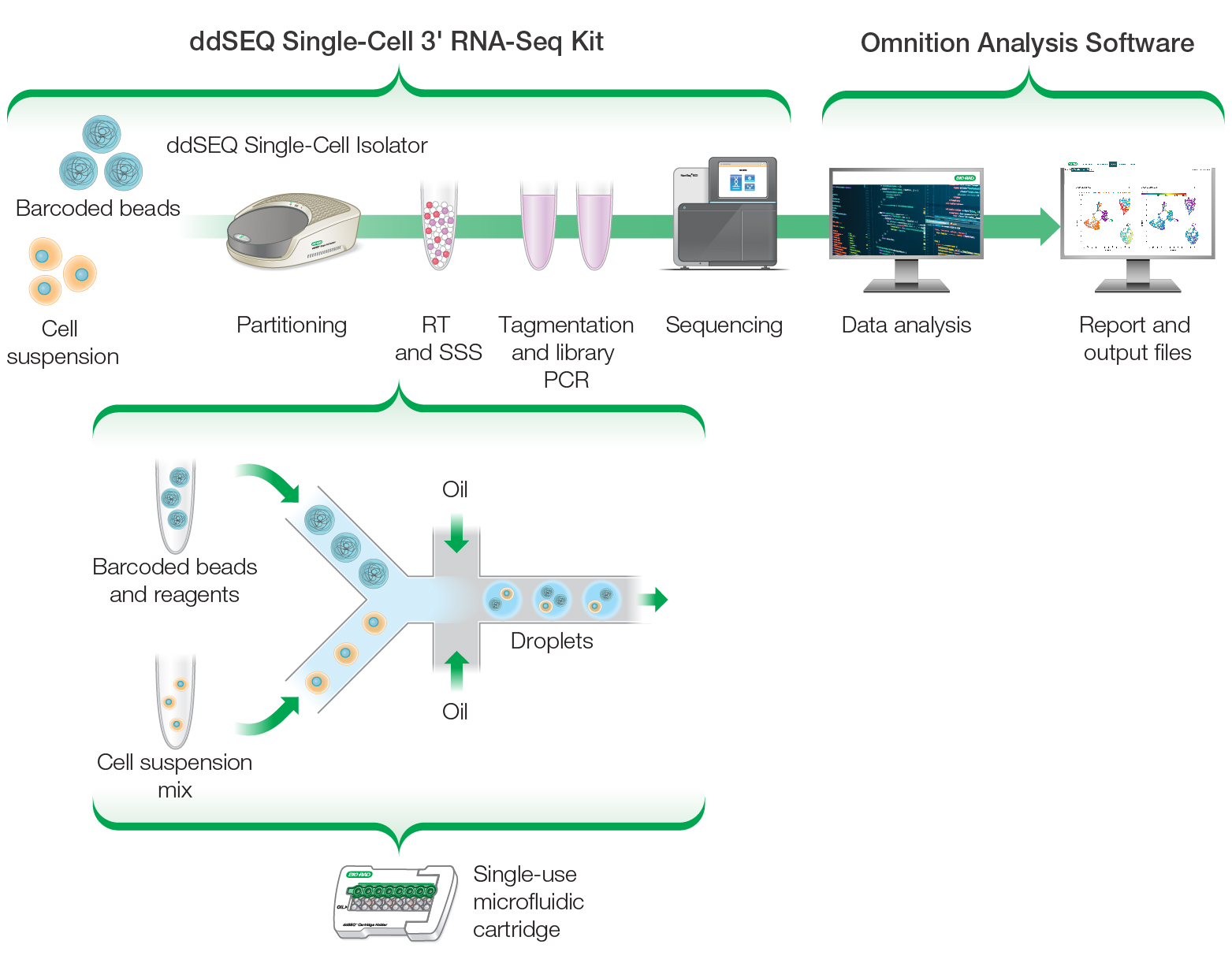
Fig. 1. The ddSEQ Single-Cell 3′ RNA-Seq Kit workflow.
Single-Cell ATAC Sequencing to Explore Chromatin Accessibility
Beyond the transcriptome, researchers can also delve into regulatory modifications affecting gene expression and cell heterogeneity in single cells by using the single-cell assay for transposase-accessible chromatin using sequencing (scATAC- Seq). Capable of assessing chromatin accessibility and the activities of regulatory elements, scATAC-Seq provides insights into the epigenetic mechanisms dictating cell behavior, with applications in biomarker discovery, T-cell activation, and cancer.
By incorporating whole genome open chromatin interrogation into single-cell workflows, scATAC-Seq can unravel the complexities of cellular heterogeneity, differentiation, and disease-specific alterations in gene regulation. In particular, it can be used to study cell-specific chromatin accessibility and characterize regulatory signatures in tissue samples with a heterogeneous cellular population. When used in combination with other techniques, such as scRNA-Seq, scATAC-Seq can be part of a multi-omics analysis workflow, offering a deeper understanding of gene regulation in cellular processes and disease.
Using the same droplet technology employed in our ddSEQ Single-Cell 3′ RNA-Seq Kit, the ddSEQ Single-Cell ATAC-Seq Library Prep Kit and ddSEQ Single-Cell Isolator provide fast (one-day workflow), cost-efficient, high-quality, and sensitive genome-wide profiling of the epigenomic landscape at the single-cell level (Figure 2).
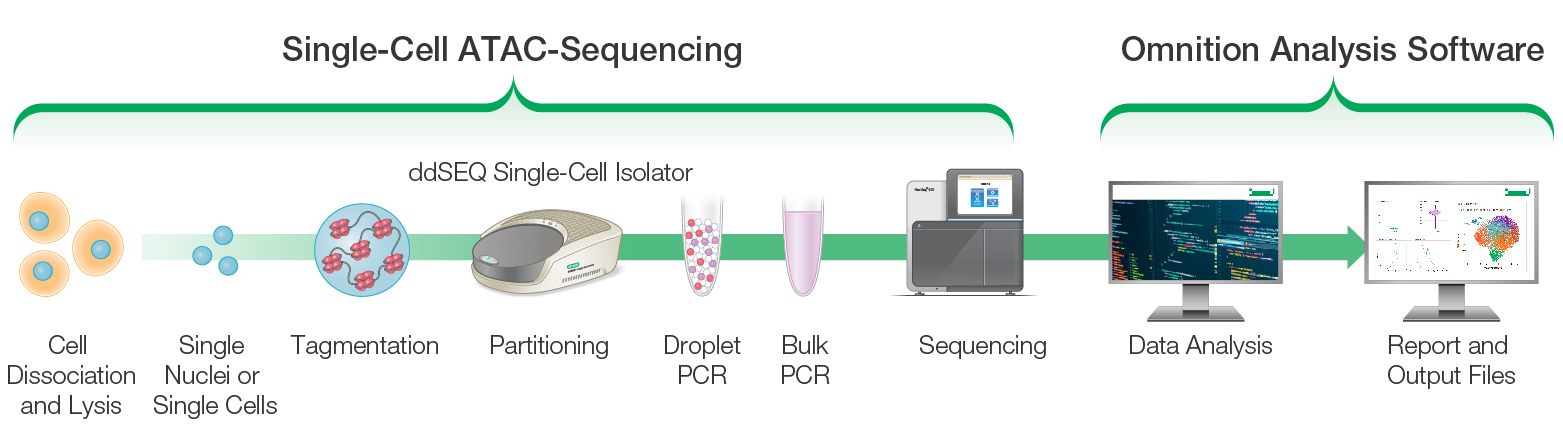
Fig. 2. The ddSEQ Single-Cell ATAC-Seq Kit workflow.
Conclusion
The Bio-Rad portfolio of RNA-Seq technologies offers simple, comprehensive workflows, enabling efficient cell isolation and streamlined mapping of the transcriptome to provide insights into disease and other biological mechanisms. The high-throughput droplet approach is highly scalable and cost-effective, reducing barriers to single-cell analysis and opening opportunities for both new and existing users, while facilitating more effective disease diagnosis and monitoring.
Visit our website to learn more about single-cell library preparation for next-generation sequencing and reach out to our experts with your questions about single-cell 3′ RNA-Seq.
References
Awuah WA et al. (2023) The molecular landscape of neurological disorders: insights from single-cell RNA sequencing in neurology and neurosurgery. Eur J Med Res 28, 529.
Choi JR et al. (2020) Single-cell RNA sequencing and its combination with protein and DNA analyses. Cells 9, 1130.
Haque A et al. (2017) A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med 9, 75.
Lim J et al. (2024) Advances in single-cell omics and multiomics for high-resolution molecular profiling. Exp Mol Med 56, 515–526.
Ma F et al. (2019) A comparison between whole transcript and 3′ RNA sequencing methods using Kapa and Lexogen library preparation methods. BMC Genomics 20, 9.
Van de Sande B et al. (2023) Applications of single-cell RNA sequencing in drug discovery and development. Nat Rev Drug Discov 22, 496–520.

