Polyacrylamide gel electrophoresis (PAGE) is a well-established technique for the separation, detection, and analysis of proteins. SDS-PAGE is used as the first step in western blotting analysis, in the second dimension separation of the 2-D analysis of proteins in proteomics studies, and as the confirmatory step in protein integrity and purity analyses after the chromatographic separation of proteins.
Visualization of proteins after separation on polyacrylamide gels is predominantly carried out using protein stains such as Coomassie Brilliant Blue (CBB). Other stains, such as silver, zinc or some fluorescent dye-based stains are also used to determine sample quality, level of separation, and protein load. The traditional staining methods are generally time consuming and preclude the use of gels in downstream applications, such as western blotting and mass spectrometry.
Stain-free technology, a technology that existed in principle since early 2000 but was only commercialized in 2010, addresses these issues and has, as a result, changed the protein separation and analysis landscape in the past few years. Stain-free technology has been adopted by many labs for its simplicity, ease of use, and, most important, its reliability, which confers validity to the results obtained in downstream applications. The number of stain-free gels has grown exponentially in the past three years.
This article focuses on the following aspects of stain-free technology:
- Theory and concepts on how protein visualization is accomplished using stain-free technology
- Advantages of stain-free technology over more traditional methods
- How stain-free technology can be used for total protein normalization
- Possible concerns about the stain-free technology
- How stain-free technology can change the way we carry out protein separation and analysis
What Is Stain-Free Technology?
Stain-free technology utilizes the ability of certain aromatic amino acid residues to undergo specific chemical reactions when exposed to UV radiation. Aromatic amino acids, such as tryptophan and tyrosine, form key intermediates, such as kynurenine, cyclic lactams, and Dopa, upon UV irradiation. Specifically, tryptophan reacts with certain trihalo compounds to produce a tryptophan adduct that emits fluorescence. The rate at which the chemical reaction produces fluorescence is enhanced with the addition of the trihalo compound compared to the rate with UV activation alone. Stain-free technology exploits this capability for the visualization and quantitation of proteins (Figure 1).
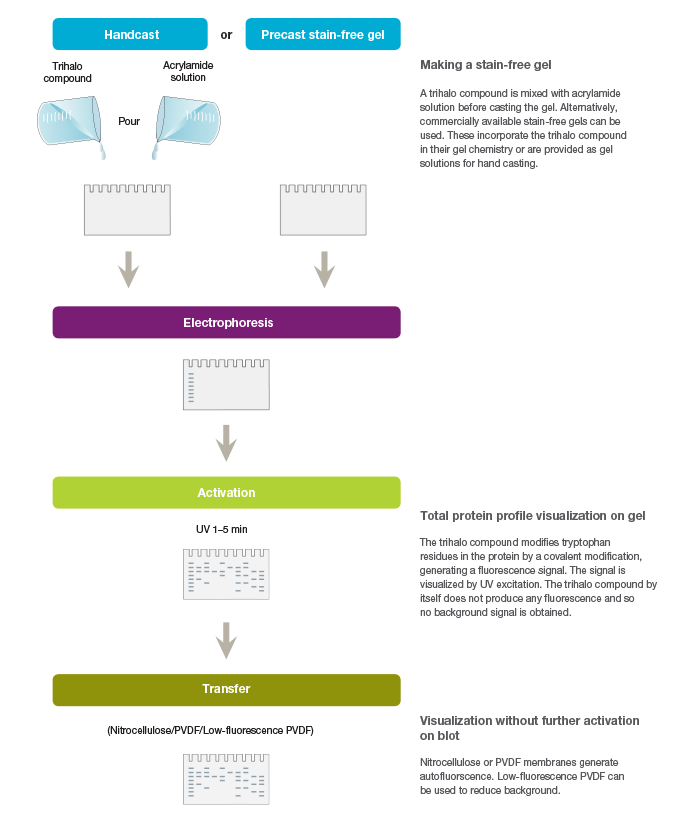
Fig. 1. Workflow for the stain-free visualization of proteins.
Commercial stain-free technology utilizes proprietary trihalo compounds contained in the gels to modify tryptophan residues in proteins following brief UV irradiation. The resulting tryptophan adduct (a 58 Da moiety) emits fluorescence upon excitation by another brief irradiation from stain-free enabled imagers, facilitating visualization and quantitation of proteins from gels, as well as from blots after the proteins are transferred or probed with antibodies for western blotting. The limit of detection for the visualization of proteins rendered by this method is 20–50 ng of protein compared to the 100 ng range of the CBB method or the 8–15 ng range of colloidal CBB (Ladner et al. 2006).
The modifications to the proteins themselves using the stain-free method are minimal and do not affect protein transfer or downstream antibody binding in western blotting, nor does the binding mimic any posttranslational modifications, which could potentially result in misinterpretation of results. However, the covalent binding of the trihalo compound is enough to provide high sensitivity protein detection. Therefore, unlike Coomassie Blue-stained gel, stain-free gels can be used for downstream applications such as western blotting (Elbaggari et al. 2008; Gilda and Gomes, 2013) or mass spectrometry (Liu et al. 2008, Susnea et al. 2013, Gonzalez-Fernandez et al. 2013), which makes the stain-free technology superior to most dye-based visualization techniques.
Stain-free gels incorporate the trihalo compound in their gel formulation and are run with standard protocols and reagents like any other gel used in SDS-PAGE. Unlike with Coomassie or other dyes, there is no destaining step, and the stain-free technology is environmentally safe and does not generate toxic, hazardous organic waste.
Advantages of Using Stain-Free Technology
Visualization of Proteins on Gels — Stain-Free Technology Provides More Sensitivity and Better Dynamic Range than Coomassie Stains
When a new technology is introduced, the primary objective of potential users is to learn how it compares to the existing method(s). Protein visualization data obtained from stain-free gels are comparable to those obtained from gels stained with other dyes. In general, the sensitivity of stain-free gels is equal to that of Coomassie-stained gels for all proteins. For proteins with higher tryptophan content, stain-free gels provide much higher sensitivity than Coomassie-stained gels (Figure 2).
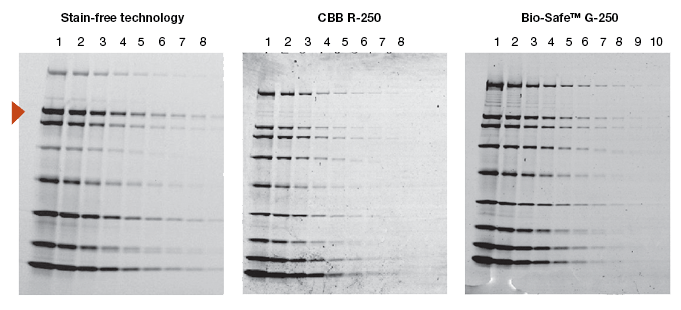
Fig. 2. Comparison of a stain-free gel image and CBB R-250– and Bio-Safe G-250– stained gel images. Serial 1:2 dilutions of broad range unstained molecular weight standards were separated on a 4–20% Criterion Stain Free™ Tris-HCl gel. The gel was imaged with a stain-free enabled imager, then stained with Coomassie stain and imaged on a densitometer. Arrowhead indicates β-galactosidase.
The limit of detection for the stain-free gels is 0.2 to 5 ng. Silver stains can provide a similar detection limit of 0.25 to 0.5 ng while Coomassie R-250 stain can detect from 6 ng of protein. Some fluorescent stains can detect proteins below 1 or 0.5 ng limit. Stain-free gels have more reproducible data with smaller coefficients of variation compared to Coomassie or silver stains. Using stain-free gels, proteins from 0.2 ng can be quantitated (McDonald et al. 2008; McDonald, 2009).
The linear dynamic range of proteins is measured as the range through which the signal intensity on a blot proportionally increases with the increase in protein load. Ideally, protein load should be such that it falls within the quantitative linear dynamic range of the antibody used for its detection (Taylor and Posch, 2014). Stain-free gels provide a linear dynamic range between 10 and 80 µg of total protein load from cell or tissue lysates at a higher range of protein load (Figure 3A), and from 20 to 1 µg at a lower range (Figure 3B) (Taylor et al. 2013, Hammond et al. 2013).
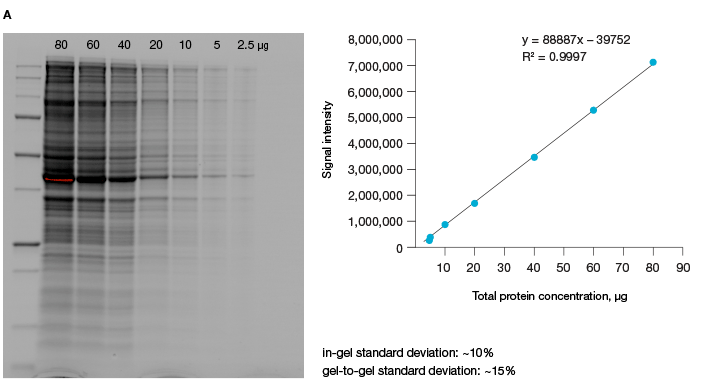
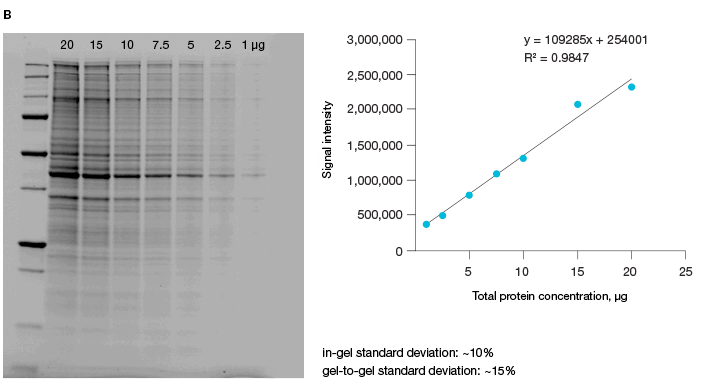
Fig 3. Linear dynamic range provided by stain-free technology for total protein measurements. A, HeLa cell lysate dilutions from 80–
2.5 µg total protein; B, HeLa cell lysate dilutions from 20–1 µg total protein.
Compatibility with Downstream Applications
The Coomassie dye binds to proteins with van der Waals and electrostatic forces, and the resulting noncovalently bound protein-dye complex emits a blue color, which is used for visualization and quantitation. This method is good as long as the goal is merely the visualization of proteins and there is no paucity of samples and reagents. The Coomassie-based staining method becomes a problem when the ultimate goal is the identification or quantitation of proteins in western blotting. This dye-based method does not allow the same gel to be used for transfer for western blotting or for mass spectrometry. This is a serious drawback, especially when large gels and sample volumes are required for separation and visualization. Stain-free technology is compatible with most downstream applications, which helps not only when resources are limited, but also in the appropriate normalization of data for quantitation (see below).
Verification of Protein Transfer
A variety of factors determine the final outcome of western blotting. One factor is the proper transfer of proteins to the membrane. A standard practice to verify protein transfer is to stain the blot with Ponceau S, which is a negatively charged stain that binds to all positively charged amino acids in a protein. Ponceau S staining is fast and relatively inexpensive, and the stained membrane can then be used for downstream applications such as western blotting. However, due to the ephemeral nature of the binding of the dye to the protein, the intensity of the bands on the membrane decreases rapidly, making detection and quantitation harder. In addition, the time needed for destaining, as well as the the need to dispose of waste, add to the disadvantages of this method. Other blot stains, such as SYPRO Ruby or amido black, are also used for blot quantitation. These are relatively expensive and need special disposal methods.
The time needed to view the stain on the blot also varies with the dye used. Protocols for using Ponceau stain recommend at least 5 minutes of staining followed by 15 minutes of washing steps. SYPRO Ruby blot stain is more elaborate, requiring fixation, overnight staining, and destaining. There is thus a significant time and cost savings for western blotting using the stain-free method (Colella et al. 2012).
Proteins transferred from a stain-free gel to a blot can be imaged using a stain-free enabled imager, and the intensity of the bands does not depend on the duration of staining or destaining, which is a problem when using dye-based techniques for visualization and quantitation (Figure 4). Band intensity of stain-free blots does not decrease with time because it is a covalent modification. Staining and destaining steps can be avoided, along with the probable protein loss during these steps. Using the stain-free method, protein transfer can be verified in as little as 2 minutes.

Fig. 4. Assessment of protein transfer using a stain-free enabled imaging system. Images of the gel before and after transfer and of the membrane after transfer were taken using the Criterion Stain Free imager. Serial 1:2 dilutions of hemoglobin (starting quantity, 80 ng), with 1.8 μg of BSA/lane as a carrier (top band), were electrophoretically separated on a 4–20% 26-well Criterion Stain Free gel.
Normalization of Data in Western Blotting Quantitation
Problems with Loading Control for Proteins and Normalization of Data
Western blotting is widely used as a semiquantitative method for the measurement of proteins. Quantitation is achieved by comparing band intensities between different samples or different experimental conditions. Protein load is an important consideration here, as misleading results can be obtained when protein loading is uneven between lanes. Loading controls that can account for simple pipetting errors are normally included in the experimental setup. Band intensity obtained from the target gene is typically normalized to that from such an internal loading control.
Traditionally, housekeeping genes, such as actin, β-tubulin, or GAPDH serve as loading controls and help in normalization of data. These specific genes are used because they are constitutively expressed and their expression is not expected to change with experimental conditions or between different tissue samples. However, there are several concerns regarding the suitability of these proteins as internal controls, arising primarily from recent findings that, contrary to popular belief, these proteins are subject to changes in different tissues (Pérez-Pérez et al. 2012), under different disease and pathological states (Li and Shen, 2013), under various experimental conditions (Dittmer and Dittmer, 2006), during various developmental stages (Rocha-Martins et al. 2012), and with different genetic backgrounds and circadian times (Kosir et al. 2010) (See Figure 5).
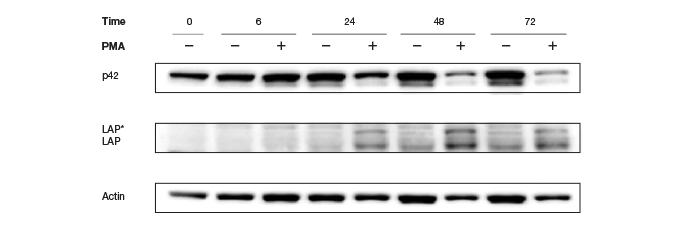
Fig. 5. Altered Actin protein expression in response to PMA treatment. Changes in expression rendered Actin an unreliable loading control.
Moreover, these controls have to be quantitated on the same gel as the protein of interest, or the same blot needs to be stripped and reprobed for the housekeeping genes, which pose their own problems and restrictions. Because of the general abundance of the housekeeping proteins relative to the proteins of interest, the intensity of the bands from a western blot of housekeeping proteins has often reached saturation (Figure 6), making normalization of data using these bands erroneous. Image quantitation software usually provides some way of selecting the bands and subtracting background. It is a good practice to obtain a standard curve for linearity of signals (intensity of bands vs. amount of protein loaded). However, this critical step is usually not part of the blot imaging protocol. Such a linear signal cannot be obtained with oversaturated bands from housekeeping proteins, making normalizing to these bands and obtaining quantitative interferences from these data prone to errors.
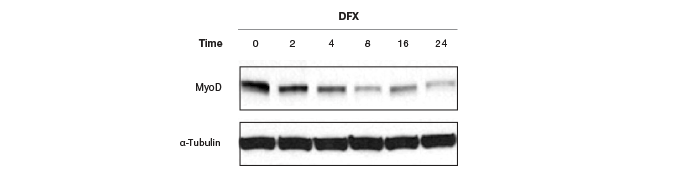
Fig. 6. Western blots showing saturated α-tubulin bands. These oversaturated bands cannot be used in this case to determine if there was an actual change in MyoD protein levels after treatment with DFX or if there was a variation in protein loading.
Stain-free technology eliminates these issues by allowing for the quantitation of total proteins on the same blot as the proteins of interest. This method of normalization of data, called total protein normalization, provides more accurate protein quantitation and western blotting results. In this method, the total density for each lane is measured from a blot and a lane profile is obtained. Specialized software from stain-free enabled imagers can interpret the data from the lanes in three dimensions, so the same lane profile data can be viewed as a three-dimensional peak. The background is adjusted in such a way that the total background is subtracted from the sum of density of all the bands in each lane (referred to as the rolling disc background subtraction algorithm; Figure 7).
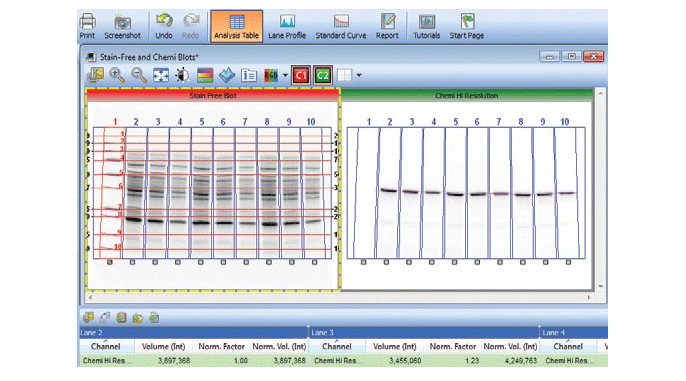
Fig. 7. Screenshot showing the software feature for total protein normalization in a stain-free enabled imager.
Normalization performed using this method is far superior to that using data from blots stripped and reprobed for housekeeping proteins. Total protein normalization using stain-free technology has also been shown to be more effective in picking up small fold differences in protein expression and regulation compared to normalization using housekeeping proteins (Gurtler et al. 2013).
The signal intensity produced from stain-free gels is much stronger (higher relative intensity) than that from Ponceau-stained membranes, and the technology provides superior linear dynamic range compared to Ponceau S total protein staining (see results from a comparative study by Gilda and Gomes 2013).
Total protein normalization also solves the issues related to the stripping and reprobing of the membranes, such as the antibody for housekeeping proteins and proteins of interest having been raised in the same organism. Despite it being a common procedure, there are several issues related to the stripping and reprobing process. An important consideration is to ensure that the primary antibody previously used is removed completely and that there is no residual signal from that particular antiserum after the membranes are stripped. This verification step, which is often skipped, requires reincubation of the blot in the ECL substrate and exposure to film before the process of reprobing begins (MacPhee 2010). Because stain-free technology does not require immunodetection of a housekeeping protein for normalization, reprobing steps can be avoided altogether.
Concerns with Using Stain-Free Technology
As stain-free technology utilizes the modification of tryptophan residues, a natural concern is the ability to use this technology for all proteins. The first and foremost concern is to determine if the protein of interest contains tryptophan residues. Proteins that lack tryptophan residues, such as aprotinin, are not detected using this technology (Figure 8). Theoretically, even one tryptophan residue is sufficient for signal activation, and proteins that have as few as two tryptophan residues are readily detected and quantified using stain-free technology.
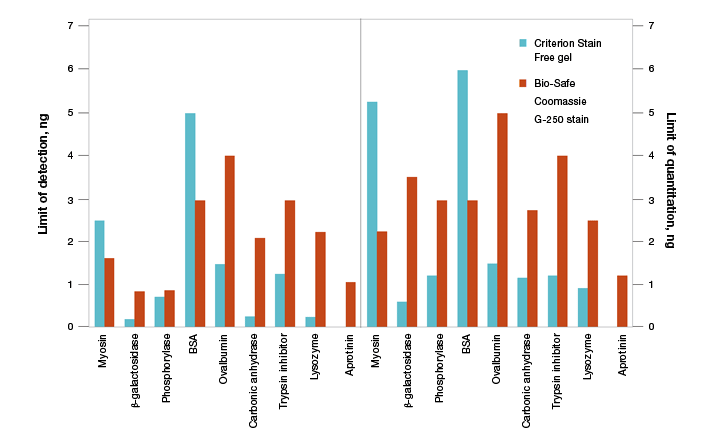
Fig. 8. Limits of detection (LOD) and limits of quantitation (LOQ) of proteins on Criterion Stain Free– and Bio-Safe G-250–stained gels. Individual protein bands from broad range unstained standards from four replicate gels were used to determine visual LOD and LOQ. Averaged numbers were used to generate the graph.
For most applications, the requirement of tryptophan residues for visualization is not a concern. Data available from UniProt (http://www.ebi.ac.uk/uniprot) show that only about 10% of proteins from all organisms lack tryptophans and most of those proteins are less than 10 kD in size. In most organisms, approximately 90% of proteins are above 10 kD in size (in the 10–260 kD range) and possess tryptophan residues (Table 1).
Table 1. Tryptophan content of the predicted proteomes of several model organisms.*

* Sequence data were obtained from UniProt (http://www.ebi.ac.uk/uniprot/database/Databases.html).
A computational study investigating the frequency of individual amino acids in the nuclear, cytoplasmic, and integral membranes of proteins in the SWISS-PROT database showed that tryptophan is present in all these fractions (Schwartz et al. 2001). Even though tryptophan is generally found less frequently than other amino acids (Shwartz et al. 2001), most proteins can be detected and quantified using the stain-free technology, with greater sensitivity and lower detection limits than with Coomassie staining (See Figure 3).
Another common concern when considering a new technology is how this technology will impact existing workflows and necessitate the use of specialized reagents and consumables. Stain-free gels do not use specialized buffers or reagents. Standard SDS-PAGE buffers can be used. However, the technique does require an imager that can visualize the activated gel or membrane.
How Is Stain-Free Technology Likely to Affect the Way We Do Protein Research Today?
With increasing errors in quantitation arising due to the use of housekeeping proteins and associated normalization issues, a growing number of journals in the western blotting field have been forced to withdraw published reports, issue errata, or reject publications (Neill 2009). Journals are now requiring strict adherence to the use of internal controls and are mandating the use of imaging techniques that yield linear signal ranges and report linear dynamic range of signal (see guidelines for reporting life sciences research from Nature Publication Group 2013).
With these new mandates, the protocols for western blotting are being rewritten. Including loading controls, checking for linearity of signal range, and ensuring proper normalization are not optional, but the norm of western blotting moving forward. Stain-free technology fits perfectly in this new western blotting world order as it satisfies all the requirements for carrying out these new protocols while making it easier than traditional methods and is considered as a strategy to improve accuracy of western blots (Ghosh et al. 2014). As more and more researchers embrace this technology, the future of western blotting and stain-free technology seem to be intertwined.
References
Colella AD et al. (2012). Comparison of stain-free gels with traditional immunoblot loading control methodology. Anal Biochem 430, 108–110.
Dittmer A and Dittmer J (2006). Beta-actin is not a reliable loading control in western blot analysis. Electrophoresis 27, 2844–2845.
Elbaggari A et al. (2008). Evaluation of the Criterion Stain Free gel imaging system for use in western blotting applications. Bio-Rad Bulletin 5781.
Ghosh R et al. 2014. The necessity of and strategies for improving confidence in the accuracy of western blots. Expert Rev Proteomics. Jul 25, 1–12. [Epub ahead of print].
Gilda J and Gomes AV. (2013). Stain-Free total protein staining is a superior loading control to β-actin for Western blots. Anal. Biochem. Sept.15, 440(2), 186–188.
Gonzalez-Fernandez R et al. (2013). Proteomic analysis of mycelium and secretome of different Botrytis cinerea wild-type strains. J Proteomics 97, 195–221.
Gurtler A et al. (2013). Stain-free technology as a normalization tool in western blot analysis. Anal Biochem 433, 105–111.
Hammond M et al. (2013). A method for greater reliability in western blot loading controls — stain-free total protein quantitation. Bio-Rad Bulletin 6360.
Kosir R et al. (2010). Determination of reference genes for circadian studies in different tissues and mouse strains. BMC Mol Biol 11, 60.
Ladner C et al. (2006). Identification of trichloroethanol visualized proteins from two-dimensional polyacrylamide gels by mass spectrometry. Anal Chem 78, 2388–2396.
Li R and Shen Y (2013). An old method facing a new challenge: re-visiting housekeeping proteins as internal reference control for neuroscience research. Life Sci, 92, 747–751.
Liu N et al. (2008). Compatibility of the Criterion Stain Free gel imaging system with mass spectrometric protein analysis. Bio-Rad Bulletin 5810.
MacPhee DJ (2010). Methodological considerations for improving western blot analysis. J Pharmacol Toxicol Methods 61, 171–177.
McDonald K (2009). Overcoming the Coomassie blues. Bio-Rad Bulletin 5939.
McDonald K et al. (2008). In-gel protein quantitation using the Criterion Stain Free™ gel imaging system. Bio-Rad bulletin 5782.
Neill US (2009). All data are not created equal. J Clin Invest 119, 424.
Pérez-Pérez R et al. (2012). Uncovering suitable reference proteins for expression studies in human adipose tissue with relevance to obesity. PLoS One 7, e30326.
Rocha-Martins M et al. (2012). Avoiding pitfalls of internal controls: validation of reference genes for analysis by qRT-PCR and western blot throughout rat retinal development. PLoS One 7, e43028.
Schwartz R et al. (2001). Whole proteome pI values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res 11, 703–709.
Susnea I et al. 2013. Application of MALDI-TOF-mass spectrometry to proteome analysis using stain-free gel electrophoresis. Top Curr Chem. 331, 37-54, doi: 10.1007/128_2012_321.
Taylor SC et al. (2013). A defined methodology for reliable quantification of western blot data. Mol Biotechnol 55, 217–226.
Taylor SC and Poshch A (2014). The design of a quantitative western blot experiment. Biomed Res Int. 2014, 361590. doi: 10.1155/2014/361590. Epub 2014 Mar 16.
Coomassie is a trademark of BASF Aktiengesellschaft.
SYPRO is a trademark of Life Technologies Corporation.

